Team:DTU-Denmark/Protein Models
From 2013.igem.org
Protein Models
Contents |
We have determined the structures of some proteins we want our mutants to express. Others could be homology modelled due to existing 3D models that had high similarity to the ones we are working with.
Mutant 1
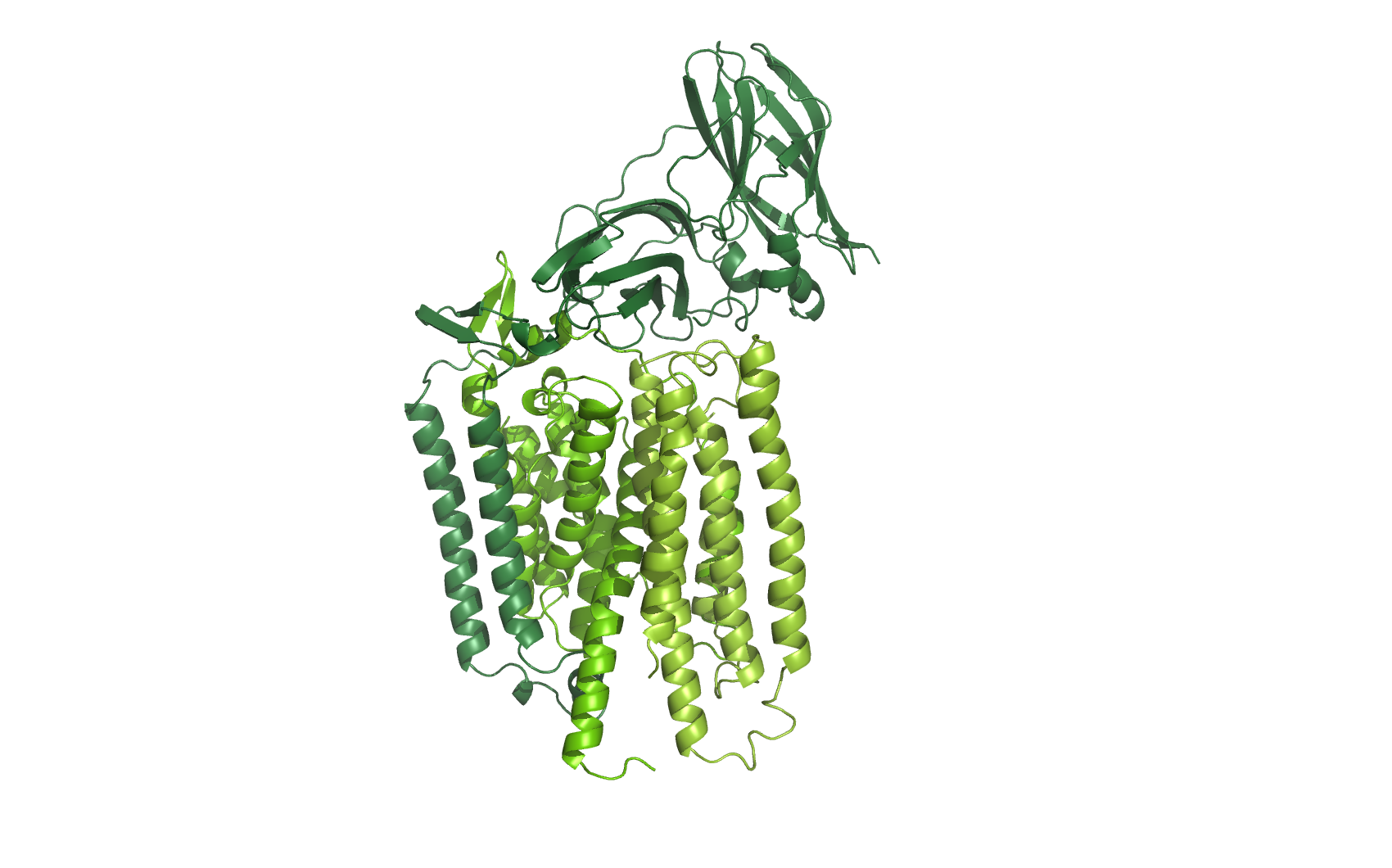
AMO
Figure 1: Ammonia monooxygenase (AMO). Homology models of all three subunits of AMO.
At the time of writing none of the three subunits from Nitrosomonas Europaea are available as 3D structures, so each of the subunits was identified at UNIPROT, and their sequence was homology modelled using CPHmodels-3.2
| Resource | AmoA | AmoB | AmoC |
|---|---|---|---|
| Uniprot | Q04507 | Q04508 | Q82T63 |
| Template pdb code and chain | 1YEW.A | 1YEW.B | 3RGB.C |
| Coverage | 90.5 | 86.2 | 66.4 |
| E-value | 1e-87 | 3e-67 | 8e-43 |
The three subunits were combined and aligned to the subunits of the Crystal structure of particulate methane monooxygenase, hence an overlap of two alpha helices.
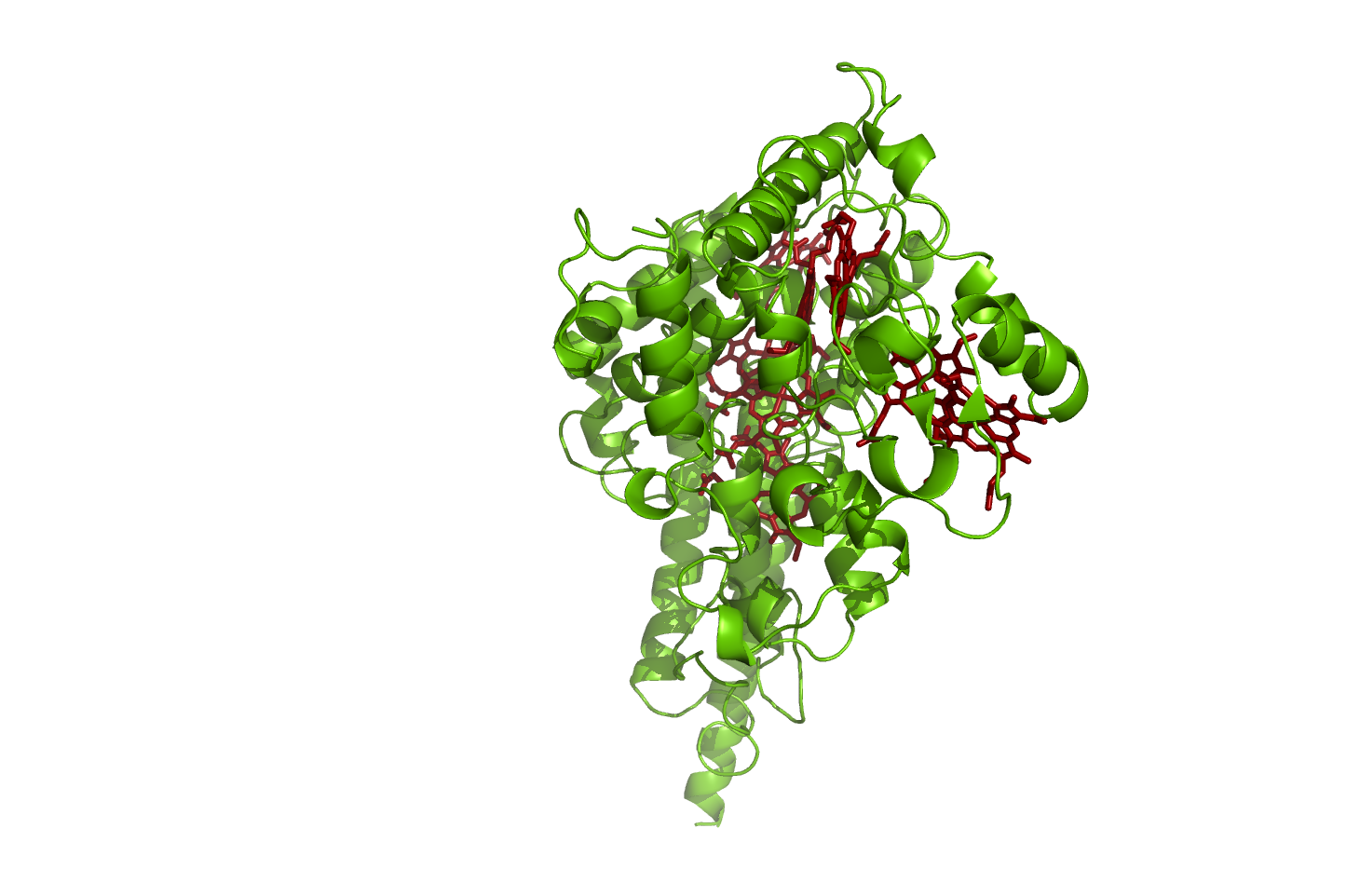
HAO
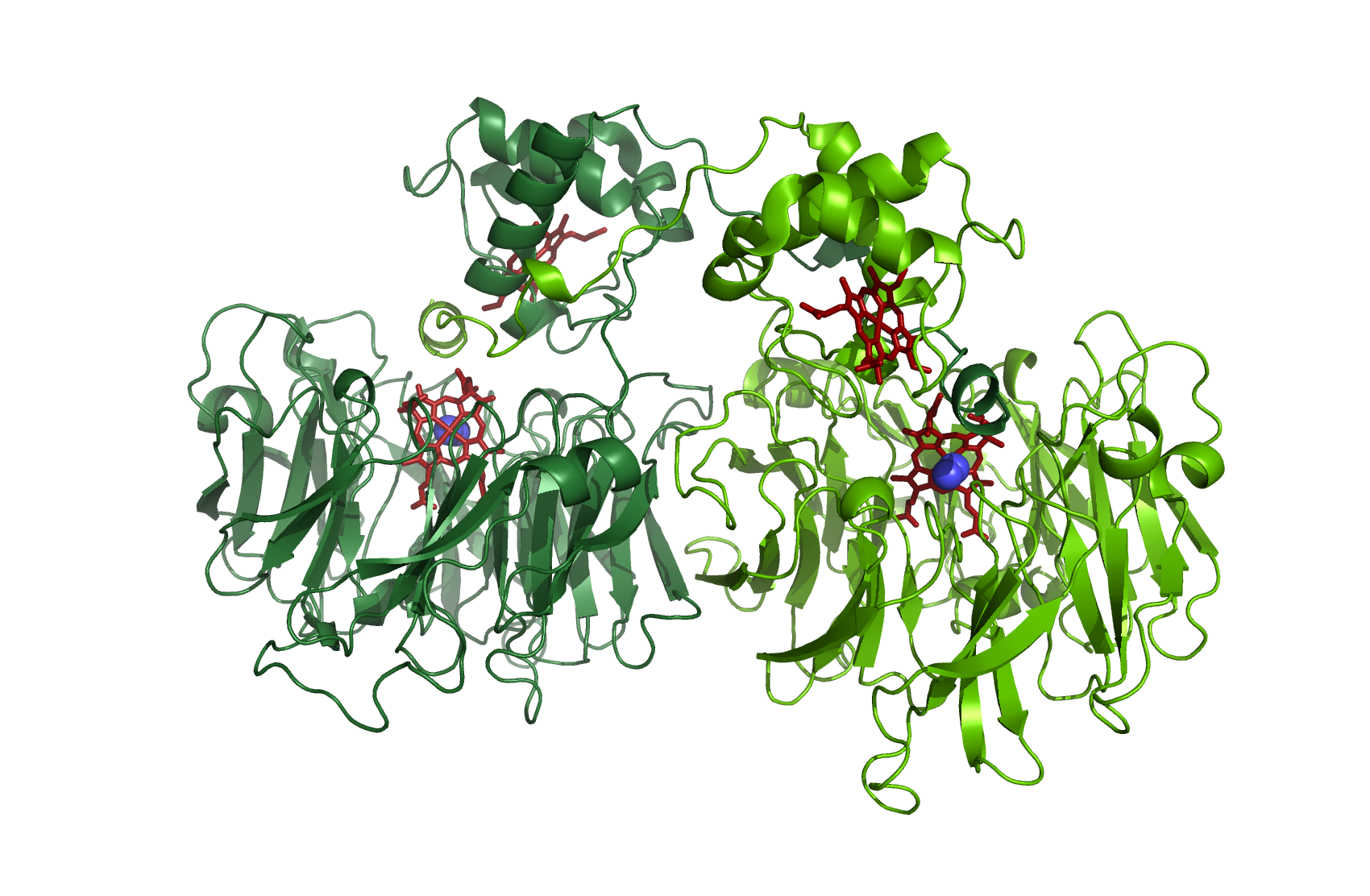
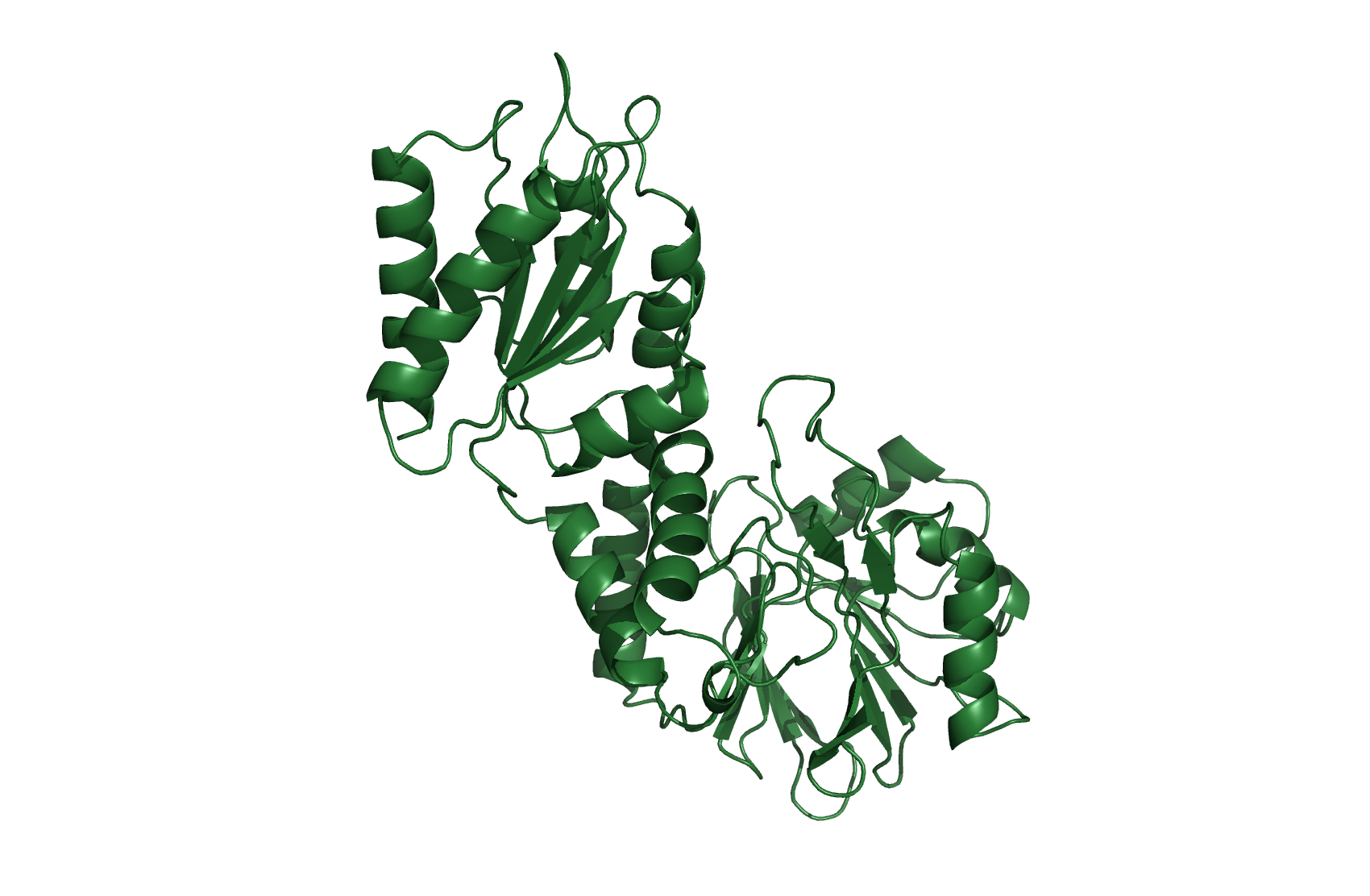
Figure 2: Hydroxylamine oxidoreductase - source 1FGJ.
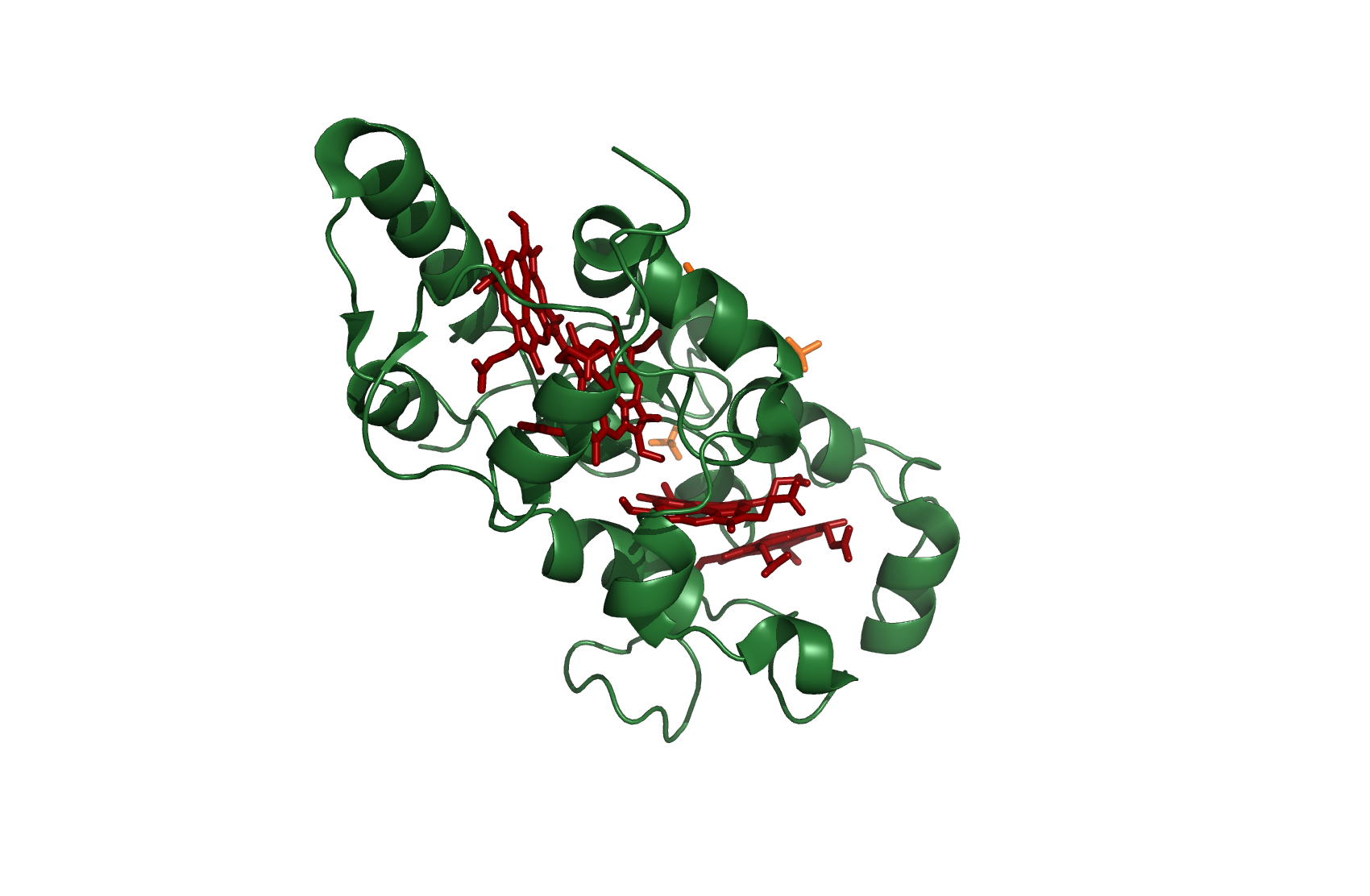
Cc554
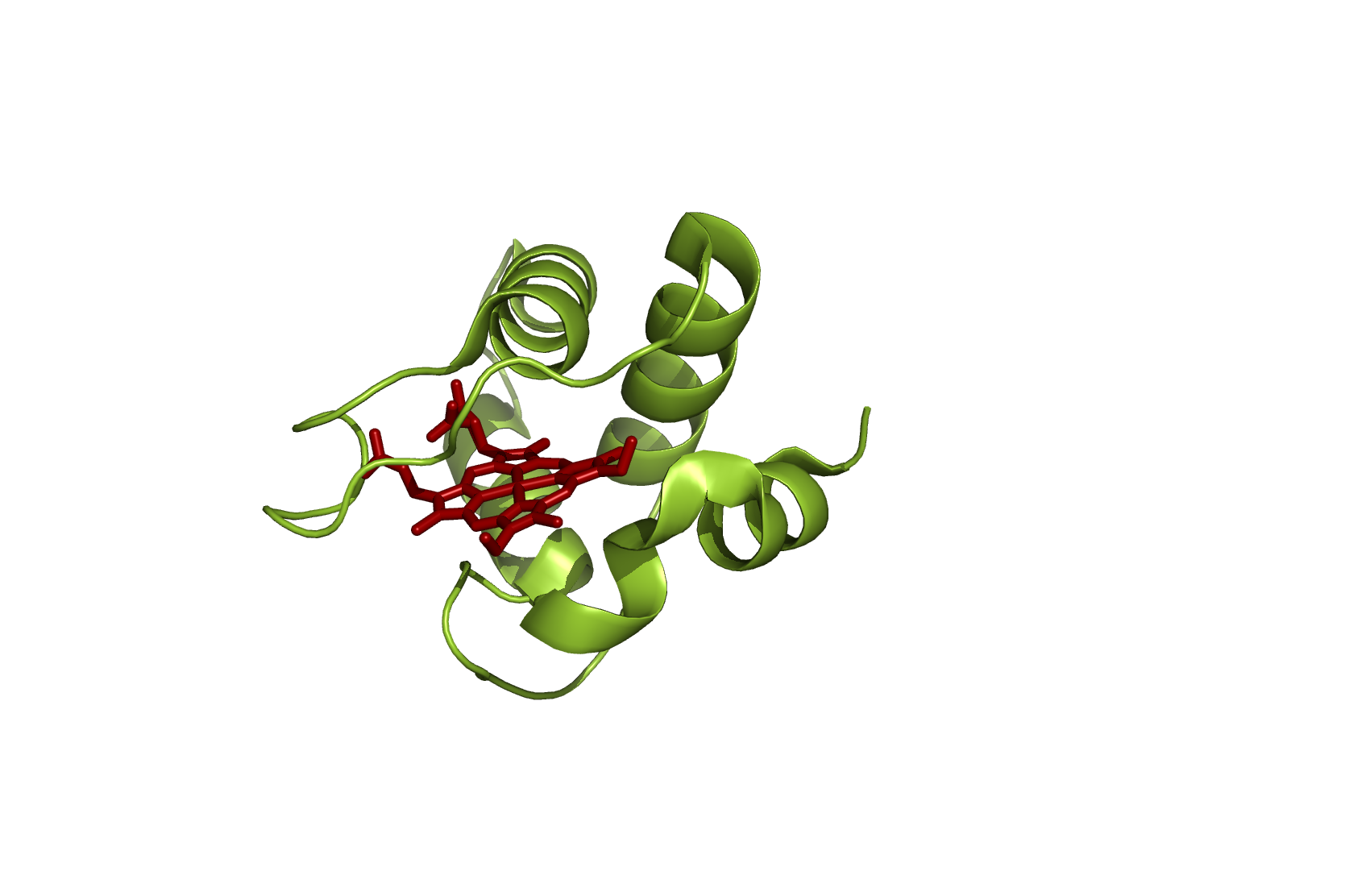
Figure 3: Cytochrome c554 - source 1FT5.
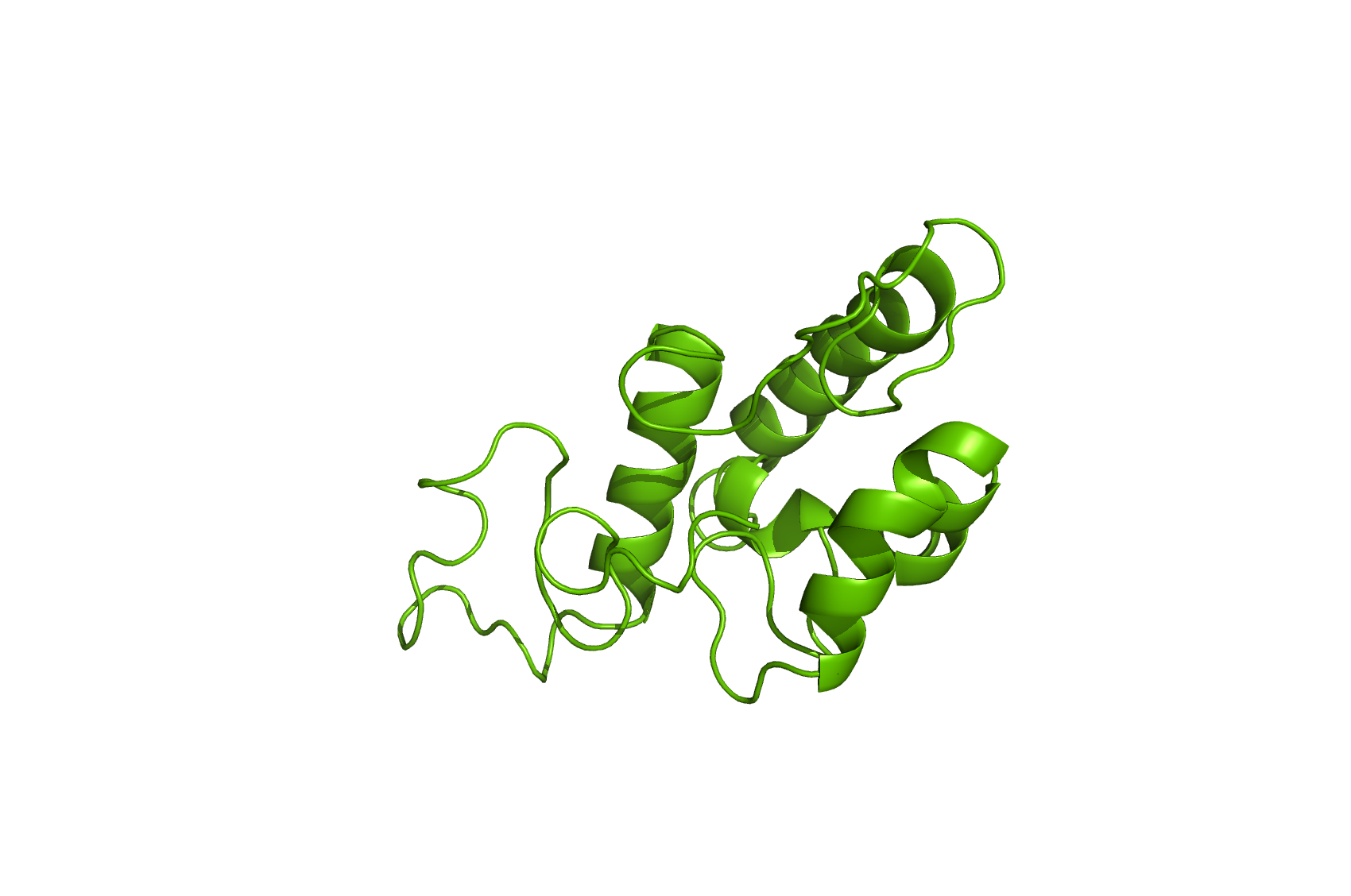
Ccm552
Figure 4: Homology model of Ccm552.
| Resource | Ccm552 |
|---|---|
| Uniprot | Q50926 |
| Template pdb code and chain | 2J7A.O |
| Coverage | 68.3 |
| E-value | 6e-11 |
Mutant 2
NirS
Figure 5: Nitrite reductase as a dimer with N2 - source 1NNO.
NirM
Figure 6: Cytochrome c551 - source 2EXV.
NOR
Figure 7: Flavorubredoxin - Homology model of NOR.
| Resource | NOR |
|---|---|
| Uniprot | Q46877 |
| Template pdb code and chain | 1YCF.A |
| Coverage | 82.5 |
| E-value | 3e-95 |
- All homology models were made using the free online tool CPHmodels v3.2
- All models were made and visualized in PyMOL v1.6 which was easily compiled and installed using an installation script made by Troels Linnet.
 "
"