Team:Frankfurt/Notebook/Labwork
From 2013.igem.org

Contents |
Labwork
Unfortunately making arrangements for the lab this year and applying for funding consumed more time than expected resulting in less time for the actual project.
July 2013
1. Arrangements for labwork such as preparing material
- see protocols for the production of competent (E.coli, S.cerevisiae) and corresponding media (LB, YEPD, SCD-ura,…)
2. Purchase/Organization of the equipment (reaction tubes, glass bottles, pipette tips,..)
August 2013
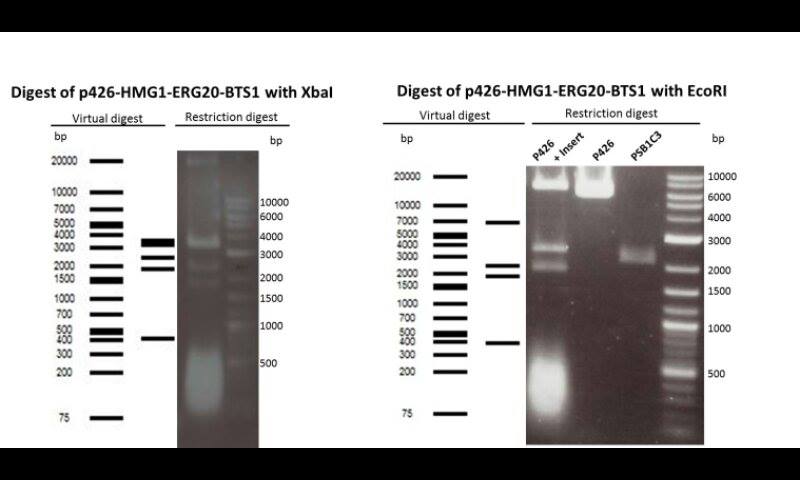
Recapitulation of the work done in 2012 : restriction digest of plasmids to make sure we can use them:
1. Digests with enzymes: BsaAI, EcoRI-HF, XbaI
example:
- 1 µl DNA (~500nm)
- 0.5 µl BsaAI (5 units / µl -> one unit cuts once within one hour)
- 1 µl Buffer 4 (10x)
- 7,5 µl ddH2O
Expectation: 4 Bands with 2246, 3196, 2344, 3624 bp. Identification of one positive clone with the EcoRI-HF digest
2. Production of competent DH5alpha
3. Digest of plasmid p426 (mevalonate) with and without insert.
4. Production of Cryo-Stocks of the p426 plasmids.
5. Transformation with the positive clone
6. Overnight cultures of yeast with the mevalonate plasmid.
7. Making cultures of p426 with and without insert for GC usage.
- Cultures were incubated for around 48 hours and harvested at an OD600 of 2.3
- Cells are centrifuged at 4000x g for 15 minutes.
September 2013
1. PCR of all fragments of the insert for the steviol plasmid (p423).
- Since pHXT7 and tCYC1 are already on the plasmid p423 they do not have to be amplified.
2. Making cultures for analysis with GC-MS
3. Gel-Extraction of following fragments: CPS-KS, KO and KAH
4. Production of comptent yeast cells (see protocols)
5. Assembly of the Steviol plasmid with Gap-repair in yeast
- yeast transformation with equimolar quantities of DNA fragments for steviol synthesis (p423 with 7 inserts)
 "
"