|
|
| Line 13: |
Line 13: |
| | | | |
| | </html> == | | </html> == |
| - | == <h2>Genaral protocol</h2> ==
| |
| - | <h3>Distribution kit</h3>
| |
| - | ----
| |
| - | <p>↓With a pipette tip, punch a hole in the foil</p>
| |
| - | <p>↓Add 10μL of dH2O,and pipetting</p>
| |
| - | <p>↓Put 5min</p>
| |
| - | <p>↓Pipette 1uL of the resuspended DNA Transformation into your desired 100μL of competent cells</p>
| |
| - | <p>↓Hold on ice for 20min</p>
| |
| - | <p>↓Heat shock at 42℃ for 30sec</p>
| |
| - | <p>↓quickly</p>
| |
| - | <p>↓On ice for 2min</p>
| |
| - | <p>↓Add 900μL of SOCborth</p>
| |
| - | <p>↓Hold at 37℃ for 30min</p>
| |
| - | <p>↓Plating 100μL of DNA Transformation</p>
| |
| - | <p>↓Centrifuge for 1 min(13,000rpm)</p>
| |
| - | <p>↓Waste supernatant for 800μL, and pipetting</p>
| |
| - | <p>↓Plating all</p>
| |
| - | <p>↓Incubate at 37℃ (over night)</p>
| |
| - | ----
| |
| - | <h3>Phenol-chloroform extraction</h3>
| |
| - | ----
| |
| - | <p>Add Phenol-chloroform where is equivalent to Exo Star process sample</p>
| |
| - | <p>↓Centrifuge 4℃ 13,000rpm 5min</p>
| |
| - | <p>Only supernatant was taken</p>
| |
| - | ----
| |
| - | <h3>EtOH crystalization</h3>
| |
| - | ----
| |
| - | <p>↓Add 1μL 20mg/mL Glycogen</p>
| |
| - | <p>↓Mix</p>
| |
| - | <p>↓Add 1/10 volume 3M CH3COONa(pH5.2)</p>
| |
| - | <p>↓Add 2.5 times volume 99.5% EtOH</p>
| |
| - | <p>↓Vortex</p>
| |
| - | <p>↓Centrifuge 4℃ 13,000rpm 20min</p>
| |
| - | <p>↓Waste supernatant</p>
| |
| - | <p>↓Add 500μL 70% EtOH</p>
| |
| - | <p>↓Mix</p>
| |
| - | <p>↓Centrifuge for 10s in a table-top microcentrifuge</p>
| |
| - | <p>↓Waste supernatant</p>
| |
| - | <p>↓65℃ Dry up</p>
| |
| - | <p>↓Add 11μL TE buffer</p>
| |
| | | | |
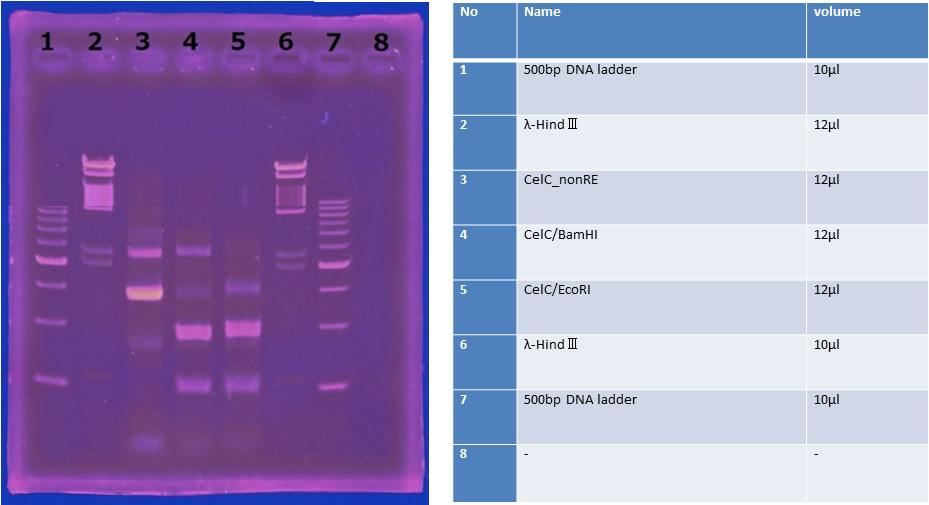
| | == <h2>CelC</h2> == | | == <h2>CelC</h2> == |
 "
"