Team:GeorgiaTech/Lambda Recombineering
From 2013.igem.org
(→Design) |
Rblackstone3 (Talk | contribs) (→Background) |
||
| Line 4: | Line 4: | ||
==Background== | ==Background== | ||
| + | Currently there is a collection of different E. coli strains called the KEIO collection, which has many different genes "knocked out" of it in order to optimize the E. coli for the proteins you wish to express. In our formation of the integrin surface unit sequence, we found through research that the deltadsbA(disulfide bond) gene would often cause to many disulfide bonds of the protein in the periplasmic space, making it impossible for the integrin to reach the surface of the cell. Also, our team used a T7 Promoter to express the integrin as this wouldbe the strongest promoter available, increasing the "decoration" of the E. coli cell with integrin surface units. To have this strain of bacteria, we needed to take the current BL21 De3 strain that creates T7 RNA Polymerase to read the vector and "knock out" the deltadsbA gene in the genome. We utilized Lambda Red Recombineering to exchange this gene. | ||
==Starting Materials== | ==Starting Materials== | ||
Revision as of 21:43, 27 September 2013
Contents |
Purpose
To replace the dsbA from BL21(De3) strain of E. coli
Background
Currently there is a collection of different E. coli strains called the KEIO collection, which has many different genes "knocked out" of it in order to optimize the E. coli for the proteins you wish to express. In our formation of the integrin surface unit sequence, we found through research that the deltadsbA(disulfide bond) gene would often cause to many disulfide bonds of the protein in the periplasmic space, making it impossible for the integrin to reach the surface of the cell. Also, our team used a T7 Promoter to express the integrin as this wouldbe the strongest promoter available, increasing the "decoration" of the E. coli cell with integrin surface units. To have this strain of bacteria, we needed to take the current BL21 De3 strain that creates T7 RNA Polymerase to read the vector and "knock out" the deltadsbA gene in the genome. We utilized Lambda Red Recombineering to exchange this gene.
Starting Materials
- Vector containing Lambda Red Recombinase (Temperature Sensitive 30C , Ampicillin Resistant)
- Electrocompetent BL21 (De3) Cells
Design
First, PKD46 needs to be transformed into an electrocompetent BL21(DE3)
This was done using electroporation
Full protocol that will be followed is shown here: File:BK-igem13-Recombineering (1).pdf
Results
July 17th, 2013
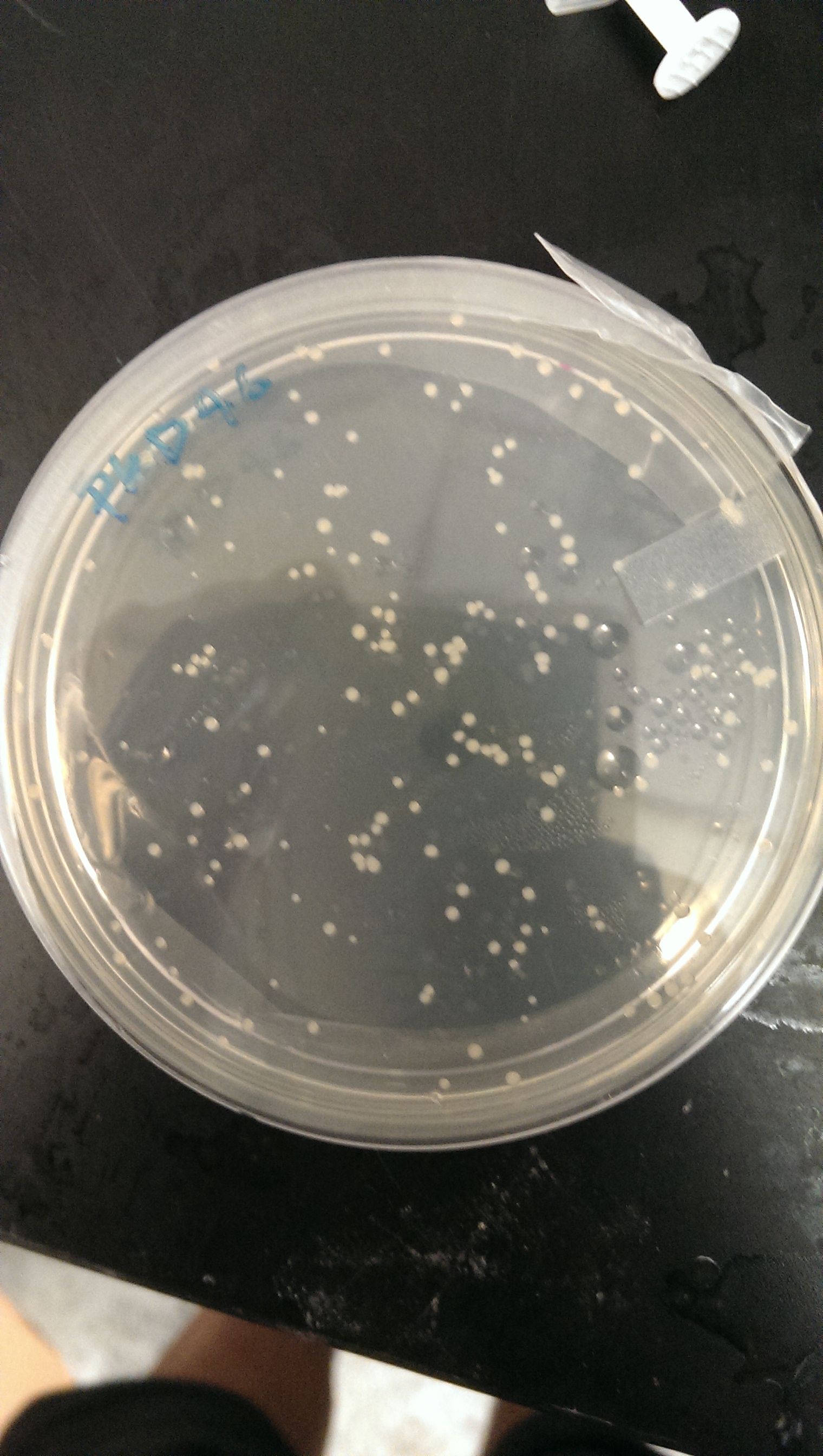
Electrocompetent transformation is done in order to transform the pkd46 vector into competent bl21(DE3) cells. The transformants were plated on an ampicillin plate.
July 18th, 2013
No growth, transformation failed.
July 19th, 2013
Transformation is repeated.
July 20th, 2013
No growth on plates yet.
July 21th, 2013
Good growth on plates. PLD46 vector has successfully been transformed into the BL21(DE3) cells.
An LB + Amp media solution is made and inoculated with a single colony
July 22th, 2013
The Bl21(DE3) strain hosting the pKD46 vector is made electrocompetent, and the dsba mutant dna (kanamycin cassette) is transformed into the cells. Cells are plated on Kanamycin + LB agar plates.
July 23th, 2013
No growth on kanamycin plates. Transformation failed
 "
"