|
|
| (25 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| - | <html xmlns="http://www.w3.org/1999/xhtml" xml:lang="en" lang="en" dir="ltr">
| + | {{:Team:Warsaw/Templates/StandardPageBegin|Acrylamide detection}} |
| - | <head>
| + | __TOC__ |
| - | <meta http-equiv="Content-Type" content="text/html; charset=utf-8" />
| + | ==Hemoglobin-based sensor== |
| - | <meta name="generator" content="MediaWiki 1.16.5" />
| + | |
| - | <link rel="alternate" type="application/x-wiki" title="Edit" href="/wiki/index.php?title=Team:Warsaw/Acrylamide_detection&action=edit" />
| + | |
| - | <link rel="edit" title="Edit" href="/wiki/index.php?title=Team:Warsaw/Acrylamide_detection&action=edit" />
| + | |
| - | <link rel="shortcut icon" href="/favicon.ico" />
| + | |
| - | <link rel="search" type="application/opensearchdescription+xml" href="/wiki/opensearch_desc.php" title="2013.igem.org (en)" />
| + | |
| - | <link title="Creative Commons" type="application/rdf+xml" href="/wiki/index.php?title=Team:Warsaw/Acrylamide_detection&action=creativecommons" rel="meta" />
| + | |
| - | <link rel="copyright" href="http://creativecommons.org/licenses/by/3.0/" />
| + | |
| - | <link rel="alternate" type="application/atom+xml" title="2013.igem.org Atom feed" href="/wiki/index.php?title=Special:RecentChanges&feed=atom" /> <title>Team:Warsaw/Acrylamide detection - 2013.igem.org</title>
| + | |
| - | <style type="text/css" media="screen, projection">/*<![CDATA[*/
| + | |
| - | @import "/wiki/skins/common/shared.css?270";
| + | |
| - | @import "/wiki/skins/igem/main.css?270";
| + | |
| - | /*]]>*/</style>
| + | |
| - | <link rel="stylesheet" type="text/css" media="print" href="/wiki/skins/common/commonPrint.css?270" />
| + | |
| - | <!--[if lt IE 5.5000]><style type="text/css">@import "/wiki/skins/igem/IE50Fixes.css?270";</style><![endif]-->
| + | |
| - | <!--[if IE 5.5000]><style type="text/css">@import "/wiki/skins/igem/IE55Fixes.css?270";</style><![endif]-->
| + | |
| - | <!--[if IE 6]><style type="text/css">@import "/wiki/skins/igem/IE60Fixes.css?270";</style><![endif]-->
| + | |
| - | <!--[if IE 7]><style type="text/css">@import "/wiki/skins/igem/IE70Fixes.css?270";</style><![endif]-->
| + | |
| - | <!--[if lt IE 7]><script type="text/javascript" src="/wiki/skins/common/IEFixes.js?270"></script>
| + | |
| - | <meta http-equiv="imagetoolbar" content="no" /><![endif]-->
| + | |
| - |
| + | |
| - | <script>
| + | |
| - | var skin="igem",
| + | |
| - | stylepath="/wiki/skins",
| + | |
| - | wgUrlProtocols="http\\:\\/\\/|https\\:\\/\\/|ftp\\:\\/\\/|irc\\:\\/\\/|gopher\\:\\/\\/|telnet\\:\\/\\/|nntp\\:\\/\\/|worldwind\\:\\/\\/|mailto\\:|news\\:|svn\\:\\/\\/",
| + | |
| - | wgArticlePath="/$1",
| + | |
| - | wgScriptPath="/wiki",
| + | |
| - | wgScriptExtension=".php",
| + | |
| - | wgScript="/wiki/index.php",
| + | |
| - | wgVariantArticlePath=false,
| + | |
| - | wgActionPaths={},
| + | |
| - | wgServer="https://2013.igem.org",
| + | |
| - | wgCanonicalNamespace="",
| + | |
| - | wgCanonicalSpecialPageName=false,
| + | |
| - | wgNamespaceNumber=0,
| + | |
| - | wgPageName="Team:Warsaw/Acrylamide_detection",
| + | |
| - | wgTitle="Team:Warsaw/Acrylamide detection",
| + | |
| - | wgAction="view",
| + | |
| - | wgArticleId=8336,
| + | |
| - | wgIsArticle=true,
| + | |
| - | wgUserName="Qba",
| + | |
| - | wgUserGroups=["*", "user", "autoconfirmed"],
| + | |
| - | wgUserLanguage="en",
| + | |
| - | wgContentLanguage="en",
| + | |
| - | wgBreakFrames=false,
| + | |
| - | wgCurRevisionId=294254,
| + | |
| - | wgVersion="1.16.5",
| + | |
| - | wgEnableAPI=true,
| + | |
| - | wgEnableWriteAPI=true,
| + | |
| - | wgSeparatorTransformTable=["", ""],
| + | |
| - | wgDigitTransformTable=["", ""],
| + | |
| - | wgMainPageTitle="Main Page",
| + | |
| - | wgFormattedNamespaces={"-2": "Media", "-1": "Special", "0": "", "1": "Talk", "2": "User", "3": "User talk", "4": "2013.igem.org", "5": "2013.igem.org talk", "6": "File", "7": "File talk", "8": "MediaWiki", "9": "MediaWiki talk", "10": "Template", "11": "Template talk", "12": "Help", "13": "Help talk", "14": "Category", "15": "Category talk"},
| + | |
| - | wgNamespaceIds={"media": -2, "special": -1, "": 0, "talk": 1, "user": 2, "user_talk": 3, "2013.igem.org": 4, "2013.igem.org_talk": 5, "file": 6, "file_talk": 7, "mediawiki": 8, "mediawiki_talk": 9, "template": 10, "template_talk": 11, "help": 12, "help_talk": 13, "category": 14, "category_talk": 15, "image": 6, "image_talk": 7},
| + | |
| - | wgSiteName="2013.igem.org",
| + | |
| - | wgCategories=[],
| + | |
| - | wgMWSuggestTemplate="https://2013.igem.org/wiki/api.php?action=opensearch\x26search={searchTerms}\x26namespace={namespaces}\x26suggest",
| + | |
| - | wgDBname="2013_igem_org",
| + | |
| - | wgSearchNamespaces=[0],
| + | |
| - | wgMWSuggestMessages=["with suggestions", "no suggestions"],
| + | |
| - | wgRestrictionEdit=[],
| + | |
| - | wgRestrictionMove=[],
| + | |
| - | wgAjaxWatch={"watchMsg": "Watch", "unwatchMsg": "Unwatch", "watchingMsg": "Watching...", "unwatchingMsg": "Unwatching...", "tooltip-ca-watchMsg": "Add this page to your watchlist", "tooltip-ca-unwatchMsg": "Remove this page from your watchlist"};
| + | |
| - | </script>
| + | |
| - | <script type="text/javascript" src="/wiki/skins/common/wikibits.js?270"><!-- wikibits js --></script>
| + | |
| - | <!-- Head Scripts -->
| + | |
| - | <script src="/wiki/skins/common/ajax.js?270"></script>
| + | |
| - | <script src="/wiki/skins/common/ajaxwatch.js?270"></script>
| + | |
| - | <script src="/wiki/skins/common/mwsuggest.js?270"></script>
| + | |
| - | <script type="text/javascript" src="/wiki/index.php?title=-&action=raw&smaxage=0&gen=js&useskin=igem"><!-- site js --></script>
| + | |
| - | <!-- jQuery Javascript -->
| + | |
| - | <script type='text/javascript' src ='/common/jquery-latest.min.js'></script>
| + | |
| - | <script type='text/javascript' src ='/common/tablesorter/jquery.tablesorter.min.js'></script>
| + | |
| - | <link rel='stylesheet' type='text/css' href='/common/tablesorter/themes/groupparts/style.css' />
| + | |
| - | <link rel='stylesheet' type='text/css' href='/common/table_styles.css' />
| + | |
| - | <link rel='stylesheet' type='text/css' href='/forum/forum_styles.css' />
| + | |
| - | <script type='text/javascript' src ='/forum/forum_scripts.js'></script>
| + | |
| - | </head>
| + | |
| | | | |
| - | <body class="mediawiki ltr ns-0 ns-subject page-Team_Warsaw_Acrylamide_detection">
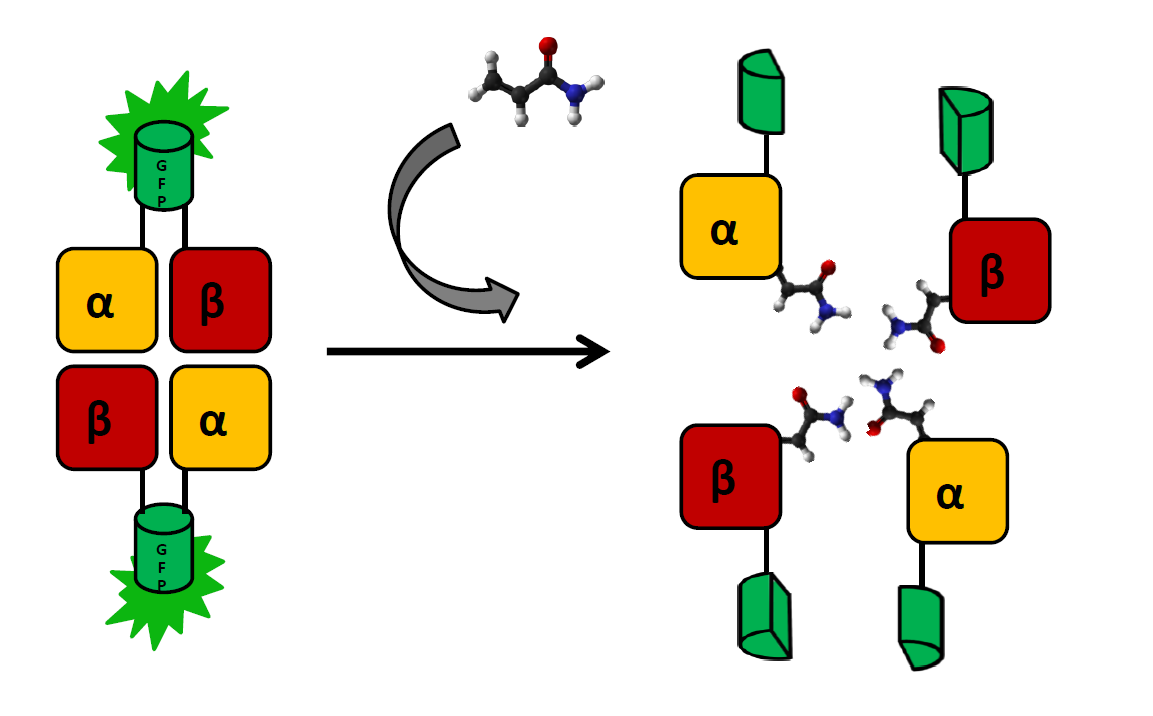
| + | The main indicator of exposure to acrylamide are adducts formed at the N-terminal valine of both hemoglobin chains. These modifications can (and have been) be detected by coating a glass electrode with human hemoglobin an measuring changes in intensity to voltage ratio (Krajewska, A. ''et al.''). We wish to create a cheaper and simpler variant of this detection system by applying ''E. coli'' strain that produces functional human hemoglobin (a modification of BactoBlood system created by Berkley 2007) with both α and β chains fused with split fluorophore fragments derived from sfGFP (superfolder GFP) protein (Zhou, J. ''et al.'') at the C-terminus. Constructs expressing each fusion protein would be transformed into two separate ''E. coli'' cultures, which would then be exposed to a solution containing acrylamide and lysed by inducing one of two kill-switches designed this year (see: Safety). |
| - | <div id="globalWrapper">
| + | |
| | | | |
| - | <div id="top-section">
| + | As the presence of acrylamide adducts at the N-terminus of hemoglobin chains results in a decrease of mutual affinity, it is also expected to lead to a significantly weaker fluorescence in relation to controls. There will be two negative controls: |
| - | <div id="p-logo">
| + | * detection system in a solution without acrylamide |
| - | <a href="/Main_Page"
| + | * bJun/bFos proteins (see: BiFC toolbox) fused with the same split fluorophore fragments and exposed to the same acrylamide solution. Interaction between those proteins should not be affected by acrylamide |
| - | title="Main Page">
| + | |
| - | <img src='/wiki/skins/common/images/wiki.jpg'>"
| + | |
| - | </a>
| + | |
| - | </div> <!-- end p-logo -->
| + | |
| - | <script type="text/javascript"> if (window.isMSIE55) fixalpha(); </script>
| + | |
| | | | |
| | + | Subunits will be expressed separately, as the formation of functional fluorophore is generally irreversible. sfGFP was chosen for this system, as superfolder mutations result in better resistance to unspecific quenching: |
| | | | |
| | | | |
| - | <div id="menubar" class='left-menu noprint'>
| + | [[File:Hemo.png|600px|thumb|center|A simplified overview of the hemoglobin-based sensor.]] |
| - | <ul>
| + | |
| - | <li
| + | |
| - | class='selected' ><a href="/Team:Warsaw/Acrylamide_detection">Page </a></li>
| + | |
| - | <li
| + | |
| - | class='new' ><a href="/wiki/index.php?title=Talk:Team:Warsaw/Acrylamide_detection&action=edit&redlink=1">Discussion </a></li>
| + | |
| - | <li
| + | |
| - | ><a href="/wiki/index.php?title=Team:Warsaw/Acrylamide_detection&action=edit">Edit </a></li>
| + | |
| - | <li
| + | |
| - | ><a href="/wiki/index.php?title=Team:Warsaw/Acrylamide_detection&action=history">History </a></li>
| + | |
| - | <li
| + | |
| - | ><a href="/Special:MovePage/Team:Warsaw/Acrylamide_detection">Move </a></li>
| + | |
| - | <li
| + | |
| - | ><a href="/wiki/index.php?title=Team:Warsaw/Acrylamide_detection&action=watch">Watch </a></li>
| + | |
| - | <li style='color:white;cursor:default'>teams</li>
| + | |
| - | </ul>
| + | |
| - | </div> <!-- end menubar (left) -->
| + | |
| | | | |
| - | <div class="right-menu noprint" id="menubar">
| |
| - | <ul>
| |
| - | <li id="pt-userpage"><a href="/User:Qba" title="Your user page [.]" accesskey="." class="new">Qba</a></li> <li id='pt-preferences'><a href='https://igem.org/User_Information' title='My account'>My account</a></li>
| |
| - | <li id="pt-logout"><a href="/wiki/index.php?title=Special:UserLogout&returnto=Team:Warsaw/Acrylamide_detection" title="Log out">Log out</a></li> </ul>
| |
| - | </div><!-- end right menubar -->
| |
| | | | |
| - | <div id="search-controls" class="noprint">
| + | ==roGFP-based sensor== |
| - | <form action="/Special:Search" id="searchform">
| + | In parralel we planned to develop a separate method of detection, based on a redox state reporter - roGFP protein. A fusion of roGFP with human glutaredoxin is a highly specifc reporter of redox potential of cellular glutathione pool (Gutscher, M. ''et al.''). Considering the fact that conjugation with glutathione (by glutathione S-tranferase) is a key pathway of detoxication following acrylamide exposure, its effect on the redox state of gluthatione pool should be easily observable (Cui, S. ''et al.''). |
| - | <input id="searchInput" name="search" type="text" title="Search 2013.igem.org [f]" accesskey="f" value="" />
| + | roGFP is a derivative of GFP with two additional cysteines introduced ''via'' site-directed mutagenesis. Depending on the redox state, they can form a disulfide bond. This leads to a shift in excitation maximum. This effect is more reliable than mere decrease in fluorescence and requires only one simple negative control. As the measurements will be conducted in ''E. coli'' cells, we designed two fusion proteins roGFP with human glutaredoxin and with ''E. coli'' glutaredoxin. Unfortunately, due to delays in our gene synthesis, we were not able to test either of the detection systems in time before wiki freeze. |
| - | <input type='submit' name="go" class="searchButton" id="searchGoButton" value="Go" title="Go to a page with this exact name if exists" />
| + | |
| - | <input type='submit' name="fulltext" class="searchButton" id="mw-searchButton" value="Search" title="Search the pages for this text" />
| + | |
| - | </form>
| + | |
| - | </div> <!-- close search controls -->
| + | |
| - | </div> <!-- close top-section-->
| + | |
| - | <div id="content">
| + | |
| - | <a name="top" id="top"></a>
| + | |
| - | <h1 class="firstHeading">Team:Warsaw/Acrylamide detection</h1>
| + | |
| - | <div id="bodyContent">
| + | |
| - | <h3 id="siteSub" class='noprint'>From 2013.igem.org</h3>
| + | |
| - | <div id="contentSub"></div>
| + | |
| - | <!--
| + | |
| - | <div id="jump-to-nav">Jump to: <a href="#column-one">navigation</a>, <a href="#searchInput">search</a></div>-->
| + | |
| - | <!-- start content -->
| + | |
| - | <p>
| + | |
| | | | |
| - | <!-- STYLESHEET -->
| |
| - | <!-- *********** -->
| |
| | | | |
| - | <link rel="stylesheet" href="https://2013.igem.org/Team:Warsaw/stylesheet?action=raw&ctype=text/css" type="text/css" />
| + | [[File:Igemtw structure.png|600px|thumb|center|A simplified overview of the hemoglobin-based sensor.]] |
| | | | |
| - | <!-- JAVASCRIPTS -->
| |
| - | <!-- Google Analytics -->
| |
| - | <script>
| |
| - | /*(function(i,s,o,g,r,a,m){i['GoogleAnalyticsObject']=r;i[r]=i[r]||function(){
| |
| - | (i[r].q=i[r].q||[]).push(arguments)},i[r].l=1*new Date();a=s.createElement(o),
| |
| - | m=s.getElementsByTagName(o)[0];a.async=1;a.src=g;m.parentNode.insertBefore(a,m)
| |
| - | })(window,document,'script','//www.google-analytics.com/analytics.js','ga');
| |
| | | | |
| - | ga('create', 'UA-42439664-1', 'igem.org');
| + | [[File:Igemtw graphscopy.png|600px|thumb|center|A simplified overview of the hemoglobin-based sensor.]] |
| - | ga('send', 'pageview');*/
| + | |
| | | | |
| - | </script>
| |
| | | | |
| - | <!-- *********** -->
| + | '''Bibliography:''' |
| | + | # Krajewska, A., Radecki, J. & Radecka, H. A Voltammetric Biosensor Based on Glassy Carbon Electrodes Modified with Single-Walled Carbon Nanotubes/Hemoglobin for Detection of Acrylamide in Water Extracts from Potato Crisps. Sensors 8, 5832–5844 (2008). |
| | + | # Berkeley_UC @ 2007.igem.org. at [https://2007.igem.org/Berkeley_UC] |
| | + | # Zhou, J., Lin, J. & Zhou, C. An improved bimolecular fluorescence complementation tool based on superfolder green fluorescent protein. Acta biochimica et … 43, 239–244 (2011). |
| | + | # Pédelacq, J.-D., Cabantous, S., Tran, T., Terwilliger, T. C. & Waldo, G. S. Engineering and characterization of a superfolder green fluorescent protein. Nature biotechnology 24, 79–88 (2006). |
| | + | # Hanson, G. T. et al. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. The Journal of biological chemistry 279, 13044–53 (2004). |
| | + | # Gutscher, M. et al. Real-time imaging of the intracellular glutathione redox potential. Nature methods 5, 553–9 (2008). |
| | + | # Cui, S., Kim, S., Jo, S. & Lee, Y. A study of in vitro scavenging reactions of acrylamide with glutathione using electrospray ionization tandem mass spectrometry. BULLETIN-KOREAN … (2005).at [http://newjournal.kcsnet.or.kr/main/j_search/j_download.htm?code=B050816] |
| | | | |
| - | <!-- ie9.js (fixes all Internet Explorer browsers older than ie9) -->
| + | {{:Team:Warsaw/Templates/StandardPageEnd}} |
| - | <!--[if lt IE 9]>
| + | |
| - | <script src="http://ie7-js.googlecode.com/svn/version/2.1(beta4)/IE9.js"></script>
| + | |
| - | <![endif]-->
| + | |
| - | | + | |
| - | <!-- jQuery Tools (slider) -->
| + | |
| - | <script src="http://cdn.jquerytools.org/1.2.5/full/jquery.tools.min.js"></script>
| + | |
| - | <script>
| + | |
| - | $(function() {
| + | |
| - | // initialize scrollable
| + | |
| - | $(".scrollable").scrollable().navigator();
| + | |
| - | });
| + | |
| - | </script>
| + | |
| - | | + | |
| - | <!-- Navigation scroll follow -->
| + | |
| - | <script type="text/javascript">
| + | |
| - | $(window).scroll(function () {
| + | |
| - | var scrollPos = $(window).scrollTop();
| + | |
| - | if (scrollPos > 110) {
| + | |
| - | $(".navigation").addClass("stickToTop");
| + | |
| - | } else {
| + | |
| - | $(".navigation").removeClass("stickToTop");
| + | |
| - | }
| + | |
| - | if (scrollPos > 180) {
| + | |
| - | $(".toc").addClass("stickBelowNavigation");
| + | |
| - | } else {
| + | |
| - | $(".toc").removeClass("stickBelowNavigation");
| + | |
| - | }
| + | |
| - | });
| + | |
| - | </script>
| + | |
| - | | + | |
| - | <!-- MathJax (LaTeX for the web) -->
| + | |
| - | <script type="text/x-mathjax-config">
| + | |
| - | MathJax.Hub.Config({tex2jax: {inlineMath: [['$','$'], ['\\(','\\)']]}});
| + | |
| - | MathJax.Hub.Config({
| + | |
| - | TeX: {
| + | |
| - | equationNumbers: { autoNumber: "AMS" }
| + | |
| - | }
| + | |
| - | });
| + | |
| - | </script>
| + | |
| - | <script type="text/javascript" src="http://cdn.mathjax.org/mathjax/latest/MathJax.js?config=TeX-AMS-MML_HTMLorMML"></script>
| + | |
| - | | + | |
| - | <!-- Sexy Drop Down (drop down navigation) -->
| + | |
| - | <script type="text/javascript">
| + | |
| - | $(document).ready(
| + | |
| - | function(){
| + | |
| - | $("ul.subnav").parent().find("> a").append("<span> </span>");
| + | |
| - | $("ul.topnav li").hover(
| + | |
| - | function() {
| + | |
| - | // Hover over
| + | |
| - | $(this).parent().find("ul.subnav").hide();
| + | |
| - | $(this).find("ul.subnav").show();
| + | |
| - | // Hover out
| + | |
| - | $(this).hover(
| + | |
| - | function() {
| + | |
| - | },
| + | |
| - | function(){
| + | |
| - | $(this).find("ul.subnav").hide();
| + | |
| - | }
| + | |
| - | );
| + | |
| - | },
| + | |
| - | function(){
| + | |
| - | $(this).find("ul.subnav").hide();
| + | |
| - | }
| + | |
| - | | + | |
| - | );
| + | |
| - | }
| + | |
| - | );
| + | |
| - | </script>
| + | |
| - | | + | |
| - | <!-- iGem wiki hacks -->
| + | |
| - | <!-- Remove all empty <p> tags -->
| + | |
| - | <script type="text/javascript">
| + | |
| - | $(document).ready(function() {
| + | |
| - | $("p").filter( function() {
| + | |
| - | return $.trim($(this).html()) == '';
| + | |
| - | }).remove();
| + | |
| - | });
| + | |
| - | </script>
| + | |
| - | | + | |
| - | <!-- Remove "team" from the menubar -->
| + | |
| - | <script type="text/javascript">
| + | |
| - | $(document).ready(function() {
| + | |
| - | $("menubar > ul > li:last-child").remove();
| + | |
| - | });
| + | |
| - | </script>
| + | |
| - | | + | |
| - | <!-- Empty heading? - Then remove it.. -->
| + | |
| - | <script type="text/javascript">
| + | |
| - | $(document).ready(function() {
| + | |
| - | if ("" == "Acrylamide detection") {
| + | |
| - | $("#heading").remove();
| + | |
| - | }
| + | |
| - | });
| + | |
| - | </script>
| + | |
| - | | + | |
| - | | + | |
| - | <!-- HTML CONTENT -->
| + | |
| - | <!-- ************ -->
| + | |
| - | | + | |
| - | <!-- Header image -->
| + | |
| - | <div id="header" >
| + | |
| - | <div class="centering" style="width: 100%;background-color: rgb(2,37,57);background-position:center; background-image: url('https://static.igem.org/mediawiki/2013/a/a6/Header.png')">
| + | |
| - | </div>
| + | |
| - | </div>
| + | |
| - | | + | |
| - | <!-- Navigation bar -->
| + | |
| - |
| + | |
| - | | + | |
| - | <div id="navigation">
| + | |
| - | <div class="navigation">
| + | |
| - | <div class="centering">
| + | |
| - | <ul class="topnav" id="topnav">
| + | |
| - | <li class="project navitem3">
| + | |
| - | <a href="/Team:Warsaw/Acrylamide_detection">Project</a>
| + | |
| - | <ul class="subnav">
| + | |
| - | <li><a href="/Team:Warsaw/Project_description">Project description</a></li>
| + | |
| - | <li><a href="/Team:Warsaw/Acrylamide_detection">Acrylamide detection</a></li>
| + | |
| - | <li><a href="/Team:Warsaw/BiFC_Toolbox">BiFC Toolbox</a></li>
| + | |
| - | <li><a href="/Team:Warsaw/Cytotoxicity">Cytotoxicity study</a></li>
| + | |
| - | <li><a href="/Team:Warsaw/Safety">Safety</a></li>
| + | |
| - | </ul>
| + | |
| - | </li>
| + | |
| - | <li class="technical_stuff navitem4">
| + | |
| - | <a href="/Team:Warsaw/Achievements">Achievements</a>
| + | |
| - | <ul class="subnav">
| + | |
| - | <li><a href="/Team:Warsaw/Results">Results</a></li>
| + | |
| - | <li><a href="/Team:Warsaw/Parts">Parts</a></li>
| + | |
| - | <li><a href="/Team:Warsaw/Cooperation">Cooperation with other iGEM Teams</a></li>
| + | |
| - | <li><a href="/Team:Warsaw/Judging">Judging</a></li>
| + | |
| - | </ul>
| + | |
| - | </li>
| + | |
| - | <li class="background navitem5">
| + | |
| - | <a href="/Team:Warsaw/Human_Practice">Human Practice</a>
| + | |
| - | <ul class="subnav">
| + | |
| - | <li><a href="/Team:Warsaw/Strategy_overview">Strategy overview</a></li>
| + | |
| - | <li><a href="/Team:Warsaw/Cooperation_with_PWN">Cooperation with PWN</a></li>
| + | |
| - | <li><a href="/Team:Warsaw/The night of Biologists">The night of Biologists</a></li>
| + | |
| - | <li><a href="/Team:Warsaw/Science_Picnic_2013">Science Picnic 2013</a></li>
| + | |
| - | <li><a href="/Team:Warsaw/Our cooperation with mass media">Our cooperation with mass media </a></li>
| + | |
| - | </ul>
| + | |
| - | </li>
| + | |
| - | <li class="navitem4">
| + | |
| - | <a href="/Team:Warsaw/Extras" title="">Extras</a>
| + | |
| - | <ul class="subnav">
| + | |
| - | <li><a href="/Team:Warsaw/Protocols">Protocols</a></li>
| + | |
| - | <li><a href="/Team:Warsaw/Genetic_lab_journal">Genetic lab journal</a></li>
| + | |
| - | <li><a href="/Team:Warsaw/Cellular_biology_lab_journal">Cellular biology lab journal</a></li>
| + | |
| - | <li><a href="/Team:Warsaw/Glossary">Project glossary</a></li>
| + | |
| - | <li><a href="/Team:Warsaw/Adventure with grants">Adventure with grants</a></li>
| + | |
| - | </ul>
| + | |
| - | </li>
| + | |
| - | <li class="team navitem6">
| + | |
| - | <a href="/Team:Warsaw/Team" title="">Team</a>
| + | |
| - | <ul class="subnav">
| + | |
| - | <li><a href="/Team:Warsaw/Team">Team</a></li>
| + | |
| - | <li><a href="/Team:Warsaw/Acknowledgments">Acknowledgments</a></li>
| + | |
| - | <li><a href="/Team:Warsaw/Sponsors">Sponsors</a></li>
| + | |
| - | </ul>
| + | |
| - | </li>
| + | |
| - | </ul>
| + | |
| - | </div>
| + | |
| - | </div>
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | <!-- Page heading -->
| + | |
| - | <div id="heading">
| + | |
| - | <div class="centering">
| + | |
| - | <h1>
| + | |
| - | | + | |
| - | Acrylamide detection
| + | |
| - | | + | |
| - | </h1>
| + | |
| - | </div>
| + | |
| - | </div>
| + | |
| - | | + | |
| - | <!-- Prepare for content -->
| + | |
| - | <div id="innercontent">
| + | |
| - | <div class="centering">
| + | |
| - | <div class="whitebox article">
| + | |
| - | | + | |
| - | </p>
| + | |
| - | <h2><span class="editsection">[<a href="/wiki/index.php?title=Team:Warsaw/Acrylamide_detection&action=edit&section=1" title="Edit section: Hemoglobin sensor">edit</a>]</span> <span class="mw-headline" id="Acrylamide_detection">Hemoglobin sensor</span></h2>
| + | |
| - | <p>This year Warsaw iGEM team is presenting to you <b>FluoSafe - a bacterial sensor for acrylamide detection</b>. As many of you know acrylamide is usually present in biochemical laboratories. But what is the significance of this organic compound really? Well, it is a neurotoxin and its carcinogenic activity has been widely proven.
| + | |
| - | </p>
| + | |
| - | <div class="thumb tleft"><div class="thumbinner" style="width:182px;"><a href="/File:Fries.jpg" class="image"><img alt="A" src="/wiki/images/thumb/4/45/Fries.jpg/180px-Fries.jpg" width="180" height="119" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Fries.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>French fries</div></div></div>
| + | |
| - | <p>Not only is it present in our work environment but also in our food. You might think that if something is not mentioned on the food label it means that that substance cannot be found in what you eat. Well - unfortunately this is not true. Acrylamide is found in chips, french fries, and anything that contains amino acids, reducing sugar and is subjected to heat. This environment is perfect for the <a href="http://en.wikipedia.org/wiki/Maillard_reaction" class="external text" rel="nofollow">Maillard reaction</a> to occur. Once this reaction has taken place, we can find acrylamide in our food.
| + | |
| - | </p>
| + | |
| - | <div class="thumb tright"><div class="thumbinner" style="width:182px;"><a href="/File:Hemoglobin.png" class="image"><img alt="A" src="/wiki/images/thumb/c/cf/Hemoglobin.png/180px-Hemoglobin.png" width="180" height="180" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Hemoglobin.png" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Hemoglobin Structure.</div></div></div>
| + | |
| - | <p>This is the reason why our project is focusing on detecting acrylamide not only in laboratories all over the world, but also in the food we eat. Our aim is also to create a sensor that can be widely used and is cheap enought to be more available for anyone who would like to use it.
| + | |
| - | </p><p>As it has been demonstrated before acrylamide reacts with the C-terminal valine of both hemoglobin subunits, creating adducts. When this reaction has occurred, it is possible to measure acrylamide level by using ion selective electrode coated with human hemoglobin and then observing the change in current intensity to voltage ratio.
| + | |
| - | </p><p>The aim of the current project is to create a bacterial sensor using an <i>E. coli</i> strain which can produce functional human hemoglobin (this idea is based on the system designed in 2007 by the Berkeley iGEM Team).
| + | |
| - | </p>
| + | |
| - | <div class="thumb tleft"><div class="thumbinner" style="width:182px;"><a href="/File:BiFC.png" class="image"><img alt="A" src="/wiki/images/thumb/d/d6/BiFC.png/180px-BiFC.png" width="180" height="92" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:BiFC.png" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>BiFC method.</div></div></div>
| + | |
| - | <p>We plan to implement <a href="http://en.wikipedia.org/wiki/BiFC" class="external text" rel="nofollow">bimolecular fluorescence complementation (BiFC)</a> to our system. In order to do this we will create a fusion protein composed of α and β hemoglobin subunits with complementary parts of split sfGFP (an improved superfolder green fluorescent protein derivative, which is often used as a marker in molecular biology). When there is no acrylamide in the environment both chains of hemoglobin form a stabile complex allowing to reconstruct a functional, fluorescent protein.
| + | |
| - | </p><p>On the other hand, formation of acrylamide adducts at the C-end of hemoglobin chains reduce their mutual affinity, so they are unable to bring the split parts of sfGFP within proximity and fluorescence complementation cannot occur. This should lead to an observable decrease in fluorescence intensity in cells exposed to acrylamide.
| + | |
| - | </p><p>We aim to create and test a variety of constructs that will allow an implementation of different BiFC variations. We are planning to use some already applied fluorescent proteins (but unavailable in the BioBrick format): eGFP, sfGFP and mCherry. Additionally, we intend to create constructs based on proteins that have not been applied yet such as: sfCFP, sfBFP, sfYFP, sfVenus i sfCerulean (improved GFP derivatives for BiFC, with additional superfolder mutations). We also intend to compare the above-mentioned combinations to the already available Venus- and Cerulean-based systems.
| + | |
| - | </p><p>Such standardized 'BiFC toolbox' (with measured sensitivity and specificity for each individual combination of fragments) will be an important contribution to the Registry of Standard Biological Parts.
| + | |
| - | </p><p>In parallel, our second goal is to develop an alternative acrylamide sensor based on redox state reporter – the roGFP protein.The roGFP protein fused with human gluteredoxin is a highly-specific sensor of the cellular glutathione redox potential (glutathione is a simple tripeptide which takes part e.g. in the cell detoxication process).
| + | |
| - | </p><p><br />
| + | |
| - | It should be emphasized that in this method the redox state change does not lead to a decrease in the level of fluorescence, but actually causes a shift in the excitation maximum, thus making the outcomes more noticeable. The typical way of acrylamide detoxication in a cell involves conjugating acrylamide to glutathione by S-glutathione transpherase (Cui. S et al.). Thus, the influence of acrylamide on cellular glutathione pool can be easily applied for detecting its presence and concentration changes in tested solution.
| + | |
| - | </p>
| + | |
| - | <div class="thumb tleft"><div class="thumbinner" style="width:182px;"><a href="/File:HeLa-cells.jpg" class="image"><img alt="A" src="/wiki/images/thumb/e/eb/HeLa-cells.jpg/180px-HeLa-cells.jpg" width="180" height="126" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:HeLa-cells.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>HeLa cells</div></div></div>
| + | |
| - | <p><br />
| + | |
| - | Our final goal is not only to construct our sensor but also to examine the cytotoxicity caused by acrylamide exposure. Our aim is to show how acrylamide may affect different tissues. This part of our project is going to demonstrate how different cell lines (i.e derived from different tissues) <a href="https://2013.igem.org/Team:Warsaw/Cytotoxicity" class="external text" rel="nofollow">react to acrylamide exposure</a>.
| + | |
| - | </p><p><br />
| + | |
| - | </p><p><br />
| + | |
| - | <b>Bibliography:</b>
| + | |
| - | </p>
| + | |
| - | <ol><li> Krajewska, A., Radecki, J. & Radecka, H. A Voltammetric Biosensor Based on Glassy Carbon Electrodes Modified with Single-Walled Carbon Nanotubes/Hemoglobin for Detection of Acrylamide in Water Extracts from Potato Crisps. Sensors 8, 5832–5844 (2008).
| + | |
| - | </li><li> UC Berkeley 2007 iGEM project (<a href="https://2007.igem.org/Berkeley_UC" class="external autonumber" rel="nofollow">[1]</a>)
| + | |
| - | </li><li> Zhou, J., Lin, J. & Zhou, C. An improved bimolecular fluorescence complementation tool based on superfolder green fluorescent protein. Acta biochimica et … 43, 239–244 (2011).
| + | |
| - | </li><li> Pédelacq, J.-D., Cabantous, S., Tran, T., Terwilliger, T. C. & Waldo, G. S. Engineering and characterization of a superfolder green fluorescent protein. Nature biotechnology 24, 79–88 (2006).
| + | |
| - | </li><li> Kodama, Y. & Hu, C.-D. Bimolecular fluorescence complementation (BiFC): a 5-year update and future perspectives. BioTechniques 53, 285–98 (2012).
| + | |
| - | </li><li> Hanson, G. T. et al. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. The Journal of biological chemistry 279, 13044–53 (2004).
| + | |
| - | </li><li> Gutscher, M. et al. Real-time imaging of the intracellular glutathione redox potential. Nature methods 5, 553–9 (2008).
| + | |
| - | </li><li> Cui, S., Kim, S., Jo, S. & Lee, Y. A study of in vitro scavenging reactions of acrylamide with glutathione using electrospray ionization tandem mass spectrometry. BULLETIN-KOREAN … (2005).at <a href="http://newjournal.kcsnet.or.kr/main/j_search/j_download.htm?code=B050816" class="external autonumber" rel="nofollow">[2]</a>
| + | |
| - | </li></ol>
| + | |
| - | <p>
| + | |
| - | </div>
| + | |
| - | </div>
| + | |
| - | </div>
| + | |
| - | <div id="footer-panel">
| + | |
| - | <div class="centering">
| + | |
| - | <div class="footerBox">
| + | |
| - | <h4>This project is part of iGEM</h4>
| + | |
| - | <p>Read more on the <a href="https://2013.igem.org/Main_Page">iGEM Main Page</a></p>
| + | |
| - | <br/>
| + | |
| - | <a href="https://2013.igem.org/Main_Page">
| + | |
| - | <img style="width:240px" src="https://static.igem.org/mediawiki/2013/f/fc/Igem_logo_warsaw_team.png">
| + | |
| - | </a>
| + | |
| - | </div>
| + | |
| - | <div class="footerBox">
| + | |
| - | <h4>Sponsors:</h4>
| + | |
| - | <p><a href="https://2013.igem.org/Team:Warsaw/Sponsors">Click here for full list of sponsors.</a></p>
| + | |
| - | <br/>
| + | |
| - | <table border="0" width="100%" cellpadding="2" cellspacing="3" style="background:transparent;padding: 0px;">
| + | |
| - | <tr>
| + | |
| - | <td><a href="http://www.nauka.gov.pl/"><img src="https://static.igem.org/mediawiki/2013/1/10/Sponsor_ministry1.png" width="200"></a></td>
| + | |
| - | <td><a href="http://www.poig.gov.pl/english/Strony/Introduction.aspx"><img src="https://static.igem.org/mediawiki/2013/f/f9/Sponsor_ig.png" width="200"></a></td>
| + | |
| - | <th rowspan="2"><a href="http://fuw.pl/"><img src="https://static.igem.org/mediawiki/2013/c/cb/Sponsor_fuw.jpg" width="150"></a></th>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td><a href="http://www.nauka.gov.pl/generacja-przyszlosci/generacja-przyszlosci.html"><img src="https://static.igem.org/mediawiki/2013/e/eb/Sponsor_gp1.jpg" width="200"></a></td>
| + | |
| - | <td><a href="http://www.funduszeeuropejskie.gov.pl/english/Strony/European_Funds.aspx"><img src="https://static.igem.org/mediawiki/2013/1/16/Sponsor_ue.png" width="200"></a></td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| - | | + | |
| - | | + | |
| - | <!--
| + | |
| - | <table border="0" width="100%" cellpadding="2" cellspacing="3" style="background:transparent;padding: 0px;">
| + | |
| - | <tr>
| + | |
| - | <td><a href="http://www.nauka.gov.pl/"><img src="https://static.igem.org/mediawiki/2013/1/10/Sponsor_ministry1.png" width="150"></a></td>
| + | |
| - | <td><a href="http://www.poig.gov.pl/english/Strony/Introduction.aspx"><img src="https://static.igem.org/mediawiki/2013/f/f9/Sponsor_ig.png" width="150"></a></td>
| + | |
| - | <td><a href="http://en.pwn.pl/index.php/about-pwn.html"><img src="https://static.igem.org/mediawiki/2013/b/bc/Sponsor_pwn.jpg" width="150"></a>
| + | |
| - |
| + | |
| - | </td>
| + | |
| - | <th rowspan="2"><a href="http://fuw.pl/"><img src="https://static.igem.org/mediawiki/2013/c/cb/Sponsor_fuw.jpg" width="150"></a></th>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td><a href="http://www.nauka.gov.pl/generacja-przyszlosci/generacja-przyszlosci.html"><img src="https://static.igem.org/mediawiki/2013/e/eb/Sponsor_gp1.jpg" width="150"></a></td>
| + | |
| - | <td><a href="http://www.funduszeeuropejskie.gov.pl/english/Strony/European_Funds.aspx"><img src="https://static.igem.org/mediawiki/2013/1/16/Sponsor_ue.png" width="150"></a></td>
| + | |
| - | <td><a href="http://oligo.pl/"><img src="https://static.igem.org/mediawiki/2013/2/21/Oligopl.gif" width="150"></a>
| + | |
| - | </td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| - | -->
| + | |
| - | | + | |
| - | <!--
| + | |
| - | <a href=""><img class="sponsorImage" src="https://static.igem.org/mediawiki/2013/1/10/Sponsor_ministry1.png" width="150"></a>
| + | |
| - | <a href=""><img class="sponsorImage" src="https://static.igem.org/mediawiki/2013/e/eb/Sponsor_gp1.jpg" width="150"></a>
| + | |
| - | <a href=""><img class="sponsorImage" src="https://static.igem.org/mediawiki/2013/b/bc/Sponsor_pwn.jpg" width="150"></a>
| + | |
| - | <a href=""><img class="sponsorImage" src="https://static.igem.org/mediawiki/2013/c/cb/Sponsor_fuw.jpg" width="150"></a>
| + | |
| - | <a href=""><img class="sponsorImage" src="https://static.igem.org/mediawiki/2013/2/21/Oligopl.gif" width="150"></a>
| + | |
| - | -->
| + | |
| - | </div>
| + | |
| - | </div>
| + | |
| - | </div>
| + | |
| - | | + | |
| - | </p>
| + | |
| - | <!--
| + | |
| - | NewPP limit report
| + | |
| - | Preprocessor node count: 31/1000000
| + | |
| - | Post-expand include size: 320/2097152 bytes
| + | |
| - | Template argument size: 38/2097152 bytes
| + | |
| - | Expensive parser function count: 0/100
| + | |
| - | -->
| + | |
| - | | + | |
| - | <!-- Saved in parser cache with key 2013_igem_org:pcache:idhash:8336-0!1!0!!en!2 and timestamp 20131004144818 -->
| + | |
| - | <div class="printfooter">
| + | |
| - | Retrieved from "<a href="https://2013.igem.org/Team:Warsaw/Acrylamide_detection">https://2013.igem.org/Team:Warsaw/Acrylamide_detection</a>"</div>
| + | |
| - | <div id="catlinks"><div id='catlinks' class='catlinks catlinks-allhidden'></div></div> <!-- end content -->
| + | |
| - | <div class="visualClear"></div>
| + | |
| - | </div>
| + | |
| - | </div>
| + | |
| - | <!-- PAGE FOOTER -- ITEMS FROM COLUMN ! HAVE BEEN MOVED HERE -- RDR -->
| + | |
| - | <div class="visualClear"></div>
| + | |
| - | <div id='footer-box' class='noprint'>
| + | |
| - | <div id="footer">
| + | |
| - | <div id="f-poweredbyico"><a href="http://www.mediawiki.org/"><img src="/wiki/skins/common/images/poweredby_mediawiki_88x31.png" height="31" width="88" alt="Powered by MediaWiki" /></a></div> <div id="f-copyrightico"><a href="http://creativecommons.org/licenses/by/3.0/"><img src="http://i.creativecommons.org/l/by/3.0/88x31.png" alt="Attribution 3.0 Unported" width="88" height="31" /></a></div> <ul id="f-list">
| + | |
| - |
| + | |
| - | | + | |
| - | <!-- Recentchanges is not handles well DEBUG -->
| + | |
| - | <li id="t-recentchanges"><a href="/Special:RecentChanges"
| + | |
| - | title='Recent changes'>Recent changes</a></li>
| + | |
| - | | + | |
| - | <li id="t-whatlinkshere"><a href="/Special:WhatLinksHere/Team:Warsaw/Acrylamide_detection"
| + | |
| - | title="List of all wiki pages that link here [j]" accesskey="j">What links here</a></li>
| + | |
| - | | + | |
| - | <li id="t-recentchangeslinked"><a href="/Special:RecentChangesLinked/Team:Warsaw/Acrylamide_detection"
| + | |
| - | title="Recent changes in pages linked from this page [k]" accesskey="k">Related changes</a></li>
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | <li id="t-upload"><a href="/Special:Upload"
| + | |
| - | title="Upload files [u]" accesskey="u">Upload file</a>
| + | |
| - | </li>
| + | |
| - | <li id="t-specialpages"><a href="/Special:SpecialPages"
| + | |
| - | title="List of all special pages [q]" accesskey="q">Special pages</a>
| + | |
| - | </li>
| + | |
| - | <li><a href='/Special:Preferences'>My preferences</a></li>
| + | |
| - | </ul>
| + | |
| - | </div> <!-- close footer -->
| + | |
| - | <div id='footer'>
| + | |
| - | <ul id="f-list">
| + | |
| - | | + | |
| - | <li id="t-print"><a href="/wiki/index.php?title=Team:Warsaw/Acrylamide_detection&printable=yes"
| + | |
| - | title="Printable version of this page [p]" accesskey="p">Printable version</a>
| + | |
| - | </li>
| + | |
| - | | + | |
| - | <li id="t-permalink"><a href="/wiki/index.php?title=Team:Warsaw/Acrylamide_detection&oldid=294254"
| + | |
| - | title="Permanent link to this revision of the page">Permanent link</a>
| + | |
| - | </li>
| + | |
| - | | + | |
| - | | + | |
| - | <li id="privacy"><a href="/2013.igem.org:Privacy_policy" title="2013.igem.org:Privacy policy">Privacy policy</a></li>
| + | |
| - | <li id="disclaimer"><a href="/2013.igem.org:General_disclaimer" title="2013.igem.org:General disclaimer">Disclaimers</a></li>
| + | |
| - | </ul>
| + | |
| - | </div> <!-- close footer -->
| + | |
| - | </div> <!-- close footer-box -->
| + | |
| - |
| + | |
| - | <script>if (window.runOnloadHook) runOnloadHook();</script>
| + | |
| - | </div>
| + | |
| - | <!-- Served in 0.163 secs. --></body>
| + | |
| - | | + | |
| - | </html>
| + | |
 "
"