Team:Edinburgh/Project/Results/Metal promoters Results
From 2013.igem.org
| Line 10: | Line 10: | ||
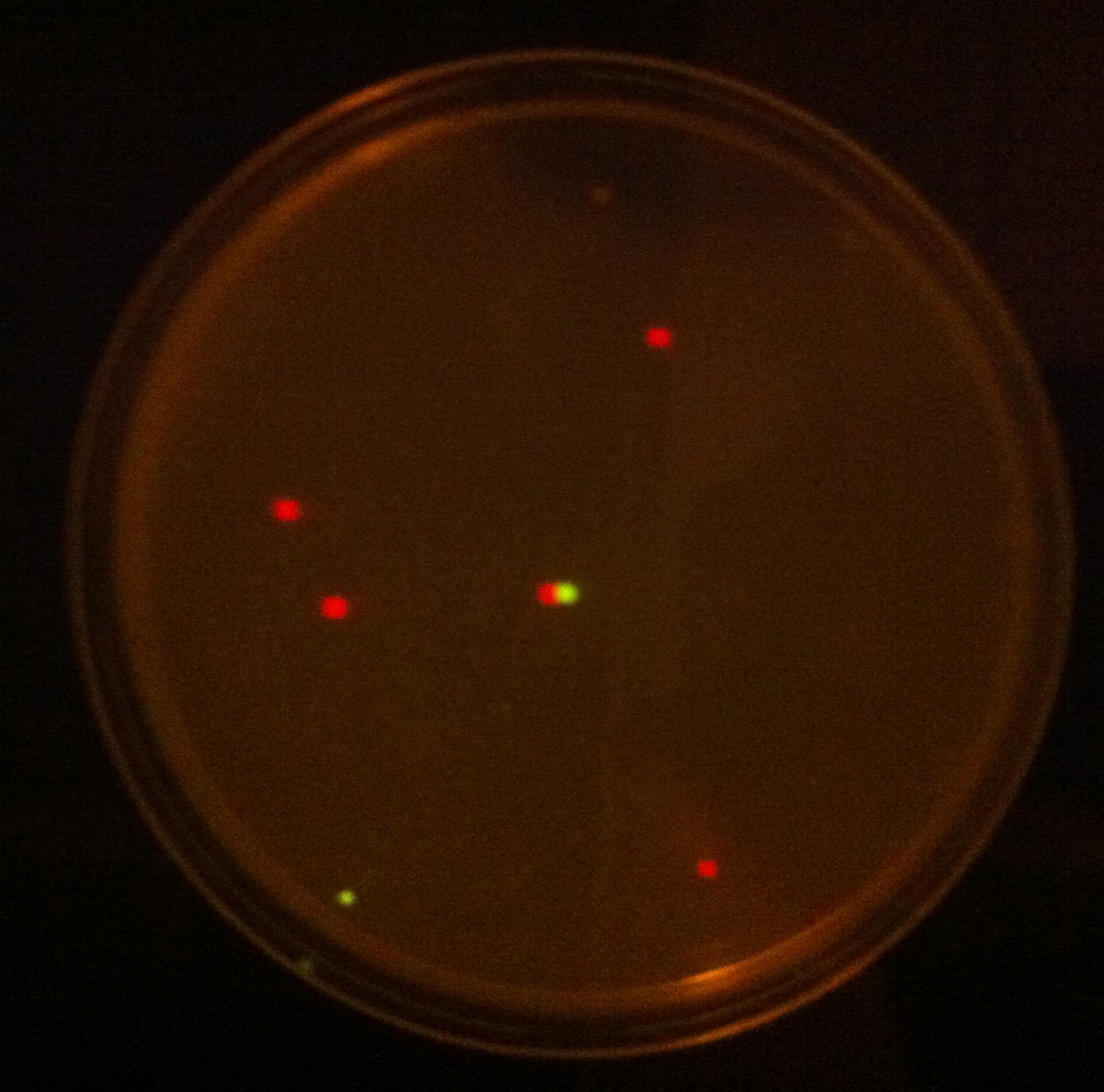
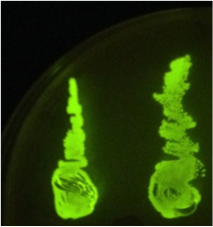
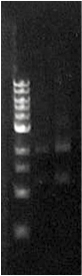
| - | When the assembly was finished, two transformations were made with competent ''E. coli'' JM109 and plated on IPTG + chloramphenicol plates and left overnight. In one plate, 4 out of 9 colonies were fluorescent; the other five had religated and were red. Figure 1 shows this plate after two GFP+ colonies were taken and restreaked; the red colonies exhibit RFP from the religated vector. The other plate had a success rate of 8 out of 20 (the competent cells were concentrated x10 before plating). Figure 2 shows the assembly fluorescing under UV when no iron was present, which proves that GFP is successfully over-expressed. Figure 3 shows the gel of restriction-digestion gel using PstI and XbaI. | + | When the assembly was finished, two transformations were made with competent ''E. coli'' JM109 and plated on IPTG + chloramphenicol plates and left overnight. In one plate, 4 out of 9 colonies were green fluorescent; the other five had religated and were red fluorescent. Figure 1 shows this plate after two GFP+ colonies were taken and restreaked; the red colonies exhibit RFP from the religated vector. The other plate had a success rate of 8 out of 20 (the competent cells were concentrated x10 before plating). Figure 2 shows the assembly fluorescing under UV when no iron was present, which proves that GFP is successfully over-expressed. Figure 3 shows the gel of restriction-digestion gel using PstI and XbaI. |
| + | |||
[[File:APlateWithRFPandGFP.jpg|600px]] | [[File:APlateWithRFPandGFP.jpg|600px]] | ||
| + | |||
| + | '''Figure 1.''' This plate has chloramphenicol and IPTG. Originally it had 4 out of 9 colonies green fluorescent and the rest red fluorescent, however two green fluorescent colonies were taken and restreaked for the iron assay. | ||
Revision as of 20:32, 4 October 2013
Fur Box Assembly
Using the protocol found in the appendix, a 3-part construct was assembled with an acceptor vector pSB1C3 (BBa_K1122009), a Lac promoter and truncated gene called PLac_LacZ (BBa_J33207) optimised for GenBrick and a fluorescent protein, GFP (BBa_K1122004). More information on the structure of the assembly can be found in the GenBrick in action section.
When the assembly was finished, two transformations were made with competent E. coli JM109 and plated on IPTG + chloramphenicol plates and left overnight. In one plate, 4 out of 9 colonies were green fluorescent; the other five had religated and were red fluorescent. Figure 1 shows this plate after two GFP+ colonies were taken and restreaked; the red colonies exhibit RFP from the religated vector. The other plate had a success rate of 8 out of 20 (the competent cells were concentrated x10 before plating). Figure 2 shows the assembly fluorescing under UV when no iron was present, which proves that GFP is successfully over-expressed. Figure 3 shows the gel of restriction-digestion gel using PstI and XbaI.
Figure 1. This plate has chloramphenicol and IPTG. Originally it had 4 out of 9 colonies green fluorescent and the rest red fluorescent, however two green fluorescent colonies were taken and restreaked for the iron assay.
|
Figure 2. Fur box Assembly. After transforming competent cells with the assembly and leaving overnight on an IPTG + chloramphenicol plate, 2 green fluorescent colonies were restreaked. A gel was performed to show the parts had correctly assembled. |
Figure 3. Fur box digest. On the left is the ladder, the first column is the undigested assembly which shows only one band (the second lower band is an artifact due to leakage). The second column contains the digested assembly. The size of PLac_LacZ + linkers + GFP was correct. |

| 
| | | | 
|
| This iGEM team has been funded by the MSD Scottish Life Sciences Fund. The opinions expressed by this iGEM team are those of the team members and do not necessarily represent those of MSD | |||||
 "
"