Team:UC-Santa Cruz/Notebook
From 2013.igem.org
2013 UCSC IGEM TEAM Polar Tags Notebook.
Groups Goals
We chose to concentrate on four polar tags found in Caulobacter Crescentus. Previous research indicated that DivJ and StpX genes were responsible for polar localization to the stalk side of the cell. Conversely PflI and TipF were chosen as candidates for polar localization to the non-stalk side or flagella side of the cell. These tags were chosen because they appear to be present in the cell during the stalked cell part of the life cycle in Caulobacter Crescentus. The stalked cells with holdfast like stalk structures are ideal for creating biofilms.
Attempts to isolate the polar tag genes from Caulobacter Crescentus involved designing PCR primers which flanked the gene of interest. Restriction sites were also added to the primers to allow for generation of fusion proteins. Generally we used Touchdown PCR cycles to attempt to amplify the polar tags out of the genome. Next the polar tags were ligated into expression vectors which contained either YFP or CFP on the C-terminal side of the multiple cloning sites. These recombinant plasmids were then transformed into competent e.coli cells for plasmid amplification. The amplified plasmids were isolated and sequenced before being transformed back into into Caulobacter.
We also identified a possible minimal (DivJM) polar tag consisting of only 235bp. This minimal DivJ was assembled using overlapping 60 mers synthesized and amplified in a PCR reaction.
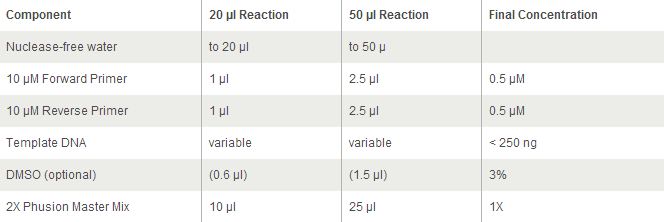
General PCR Protocol
For Phusion High-Fidelity PCR Master Mix with GC Buffer:
Initial denaturation:
98°C 30 seconds
25–35 cycles:
98°C 5–10 seconds
45–72°C 10–30 seconds
72°C 15–30 seconds/kb
Final extension:
72°C 5–10 minutes
Hold:
4°C
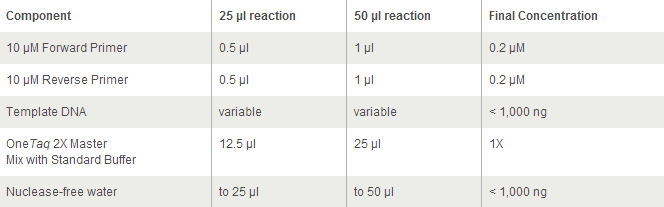
For OneTaq 2X Master Mix with Standard Buffer:
94°C 30 seconds
30 cycles:
94°C 15–30 seconds
45–68°C 15–60 seconds
68°C 1 minute per kb
Final extension:
68°C 5–10 minutes
Hold:
4-10°C
General Agarose Gel Protocol
Prepare 60 ml of 0.8% agarose solution in TBE buffer by weighing out 0.48 g of SeaKem LE agarose into an Erlenmeyer flask. Add 60 ml of TBE buffer and melt the agarose solution in the microwave. Let the agarose solution cool and pour it into the casting tray and let the agar solidify. Remove comb from the agar. Remove the casting tray from the gel box. Place the gel tray with the gel back into the gel box such that the ends of the gel are pointed into the buffer reservoirs on either end and the end of the gel with wells is pointed towards the negative electrode. Add TBE buffer to the gel box until both reservoirs are full and a 1-2 mm layer of TBE buffer covers the top (flat side facing up) side of the gel. Load samples and DNA ladder on the gel. Slide the lid and connect the electrodes to the power supply. Run the gel for 1 hour at 120 V. After gel run, take a photograph using the gel documentation apparatus.
General Chemicompetent Cell Transformation Protocol
Heat Shock Transformations:
1) Add 100 ul of Top10 Chemicompetent cells in 10 ul of each ligation reaction.
2) Place in a 42°C water bath for 30 seconds.
3) Place back on ice for 5 minutes.
4) Add 200 ul of LB to a microcentrifuge tube.
5) Transfer the Top10 Chemicompetent cells and ligation reaction to the microcentrifuge tube.
6) Incubate for one hour at 37°C at 200 rpm.
LB Media Recipe
Making LB+Kanamycin and LB+Gentamicin Agar Plates:
To make a 500 ml batch:
- 475 ml of MilliQ water
- 5 g of Tryptone
- 5 g of NaCl
- 2.5 g of yeast Extract
- 7.5 g of Agar
We made x2 500 ml batches.
Combine the reagents and shake until the solutes have dissolved. Adjust the ph to 7.0. Sterilize by autoclaving for 20 minutes on liquid cycle.
Take out of the autoclave and let the solution cool. Place in a water bath to prevent solid formation.
Add 1.25 ml of gentamicin antibiotic to one of the 500 ml batches and 500 ul of kanamycin to the other batch.
Pour into petri dishes about half way. Let agar solidify. Place in the refrigerator.
Colony Pick Protocol
Pick a colony using a pipet tip. Place the picked colony in 20 ul of MilliQ water. Pipet up and down to mix the colony thoroughly.
Electroporation Protocol
1) Obtain electrocompetent cells
2) Place sterile electroporator cuvette on ice
3) Turn on the Electroporator and adjust voltage to 1800 V
4) Add 1 ul of the ligation reaction into an aliquot of the electrocompetent cells
5) Electroporate DNA into cells. You want a time constant between 3-5 msec.
6) Pipet 1 ml of LB into cuvette pipeting up and down and transfer back into the 1.5 ml eppendorf microcentrifuge tube.
Reference Sequences
DivJ(full)
GAGCTCCTGCGACAGGATTTCCGGAGCAAGCCGATGACGGCGTTCAGAGGGGTTCGCAGTTCGTGGCTGACCATCCGCAGGAAGGAGCGCTTGCGCTCTTCCAGCGCCAGTTGGCGGCGCGCCTGTGCGGCCGCGCGGCGATCGGCCGGCGAGGGCGTGACGGCGCTGTCTTGCGACCGCGTCCTGGATCGGCCGGCTGTGCCATGGGGTCTCCCGGACCTTCTGTCCGTGCTGAATCAGATGATCAGCATGGGAGCAGGCGCTTACGATTCCGTTTCCGTCCACAGGGCTTCCACCACGAAAGGTCGCCCGCAGATTGTGCGGGGTTGGAGGGAAGGCCCGCCCCGCGTTAAGGTTTGACGACGGTGATCCTCCCCACCGCGCTAAAAAGTCGACTGGCCTTGGAATTCGAAACGCTTCCAGACCCGTTCAGACGTCCGGCCGCACGCGCCGCCGGGCTCGATCCCGCGCACGCCTGGCGGCTCGGGTGGCTGGCGGCTGTCTGTCTGGCGGCGGCGGCGGCCCTGTTCACCGCCGACTCCGGCGGTTGGCCTGTGTGGGCGGCGCTCGGCGCCGGCGCCCTGCCCGCCTTGGTCTCGCTGATCTTCACGCGGGAGGACGAACGCACCCAGTCCTGGCTTTTGGTGCTGTGGGCCGTGGGCGGCTCGCTGGCGGCGGTGCTGACCGGCGGCGTGGGCGGGGCCATGGCCGCCTGGTGTCTCGCGCCCGTGGCGGCCGCCTCCACTCAGGATCAGCCCAAGCGTCTGGCTGAGGGCGCGGCCCTGGCCCTGATCGGCGCCTGTGTCGCGGCGCTGACCCAGCTTTCCGGCCTGGCGCCCGCGGCGCCGACGGGACCGCTGGCCTTCGTTCTGGGCTTTCTGGCGCTGGTGACGACCGGTCTTGGTCTCGCCGCCGGCCTCCTGATCGGCCGGCGTCGCCAGGGCGCGCGCGACGATCGCTACGCCAGCGAGATCATCGGTCTGGAGACCTTGCTCGACGGCCTGCCGCACCTGGCGATCGCCGTCCGGGGGCAGGGACAGGTGACCGCCGTGCGCGGCGCGGCGCCGCCCGGCGTCACCCGCGCTGATCTCGTCAATCGCGGTCTGACCGGCGCGGCTGCGCCCGGCGACCGTCAGCGCCTGACCGCCGCTATCGCCCAAGCTCATCGTGAGGGCTCGGCCAGCCTGACCTTCAACCCCGCGCTGGGCGTCGAGCGCGTGGTGGCCCTGGACATGCACCGCGTTGCGCCGAACCAGCTGGTCGGCGTGCTGCGCGACATCACGGTGGAGCGGCATCGCGAGCATGCCCTCGACCAGGCGCGCATCGACGCCGAGGCCTTGGCCGCCGGCCGCGCGCGGTTCCTAGCCAATATGAGCCATGAGCTTCGCACGCCCCTGAACGCCATCATGGGCTTCTCCGACATCATGCGGGCGCGGATGTTTGGTCCGCTGAGCGACCGCTACGCCGAGTACGCTGAGCTGATCCACGAGAGCGGCGGCCATCTGCTGGACCTGATCAACGACGTGCTGGACATGTCCAAGATCGAGGCCGAGCGGTTCGAGCTGCAGCGCGGCGTGTTCGACGCGCGCGAGGCGGTTCAGGCGGCGATGCGGCTGCTGCGCGTTCAGTCTGACACCGCCGGAGTTCAGCTGCGCGGCGTCCTGCCGCCCGGCGAGCTGGAGGTCGACGCCGATCGCCGCGCGCTGAAGCAGATCGTCCTGAACCTCGTCTCGAACGCCCTGAAGTTCACCCCGCGCGGCGGGCAGGTGACCGTCACGGCGCACGGCTATGACGGCGTGCTCGAGATCGTGGTCGCCGATACGGGCGTCGGGATCAGTCCTGAGGACCTGGAACGCCTGGGGCGTCCCTACGAGCAGGCCGGCGGGGCCGAACAGCGCGCGCGGGGCACGGGCCTTGGCCTGTCGCTGGTGCGGGCTTTCGCCCAGCTGCTAGGCGGCGAGATGGTGATCGAGAGCCGCCTGGGCGCAGGCACGACCGTGTCGGTGCGCCTGCCTGTGCTGCTGGCGCCGATGGTCGCTGCCACGCCGACGCCGCCAGCCGCGCCAGAAGCGCCGTCGGCGCCGGAACCCGCTCCCACGGTTGAGGAACCGCCGCCGGCCAGCCTGGGCGACAACGTCATCGCCTTTGCGCCGCGCTGAGGCCGCTGGTCTTACCGCAGAACGCCCGACCGGCGGCGAAGTCTCCGACCTCGATATAGGCCTTGCGCGTGACCTCCTCGCCGCGTCCGGCGAAGGCGAACTCCACCGTGACCGCTTCACGAGGGTCCAGTTGCGACATCGCCTCGGCCACCGCCTCCGGAAACCGGAAGGCCAGGCCCGTCTTGGCGCCGGTCGGCAGCAGGTGACCGGGGCCTCCTCGCGCCGCTCGGCCG
DivJ326
GTGATCCTCCCCACCGCGCTAAAAAGTCGACTGGCCTTGGAATTCGAAACGCTTCCAGACCCGTTCAGACGTCCGGCCGCACGCGCCGCCGGGCTCGATCCCGCGCACGCCTGGCGGCTCGGGTGGCTGGCGGCTGTCTGTCTGGCGGCGGCGGCGGCCCTGTTCACCGCCGACTCCGGCGGTTGGCCTGTGTGGGCGGCGCTCGGCGCCGGCGCCCTGCCCGCCTTGGTCTCGCTGATCTTCACGCGGGAGGACGAACGCACCCAGTCCTGGCTTTTGGTGCTGTGGGCCGTGGGCGGCTCGCTGGCGGCGGTGCTGACCGGCGGCGTGGGCGGGGCCATGGCCGCCTGGTGTCTCGCGCCCGTGGCGGCCGCCTCCACTCAGGATCAGCCCAAGCGTCTGGCTGAGGGCGCGGCCCTGGCCCTGATCGGCGCCTGTGTCGCGGCGCTGACCCAGCTTTCCGGCCTGGCGCCCGCGGCGCCGACGGGACCGCTGGCCTTCGTTCTGGGCTTTCTGGCGCTGGTGACGACCGGTCTTGGTCTCGCCGCCGGCCTCCTGATCGGCCGGCGTCGCCAGGGCGCGCGCGACGATCGCTACGCCAGCGAGATCATCGGTCTGGAGACCTTGCTCGACGGCCTGCCGCACCTGGCGATCGCCGTCCGGGGGCAGGGACAGGTGACCGCCGTGCGCGGCGCGGCGCCGCCCGGCGTCACCCGCGCTGATCTCGTCAATCGCGGTCTGACCGGCGCGGCTGCGCCCGGCGACCGTCAGCGCCTGACCGCCGCTATCGCCCAAGCTCATCGTGAGGGCTCGGCCAGCCTGACCTTCAACCCCGCGCTGGGCGTCGAGCGCGTGGTGGCCCTGGACATGCACCGCGTTGCGCCGAACCAGCTGGTCGGCGTGCTGCGCGACATCACGGTGGAGCGGCATCGCGAGCATGCGCTCGACCAGGCGCGCATCGACGCCGAGGCCTTGGCC
DivJM
GCGGCTGCGCCCGGCGACCGTCAGCGCCTGACCGCCGCTATCGCCCAAGCTCATCGTGAGGGCTCGGCCAGCCTGACCTTCAACCCCGCGCTGGGCGTCGAGCGCGTGGTGGCCCTGGACATGCACCGCGTTGCGCCGAACCAGCTGGTCGGCGTGCTGCGCGACATCACGGTGGAGCGGCATCGCGAGCATGCGCTCGACCAGGCGCGCATCGACGCCGAGGCCTTGGCC
StpX
ATGTTTGGACGTAATATACGCTTGGCCGCCTTGGCGACCGCCGCGGCAGGCGTTCTTGCGCTGGCGGGCTGTAGTGCGGGCGATCCGGTCGCCTCGGCGCCGCCACCAAGCGATCACCAGAAATCGTCGTCGCCGCCGGAGCTGATGGGCGCGCCGCCGCCCAGCACCGCCCCAGCGCAGGACGGACTTTTGGGCGGGCCGGTCGCTGACGCCTCGGCGCAGGCCCAGGCTCAGGCGAAAGCCGACGCGGCGCGCCCGCCGCCCCCGTCGCATAACAGCCACCTGAAGACCTGGCGCCGTCCGGACGGCACCCTGGTCACGGCGATGCGGCCGATCCCCAATCCCAAGAGCGCGCCGCACGCCGCCTCGTCGGCGCCCAAGCGCGTCCACGCCAAGGTCCAGACGCAGGGCTCCTCGGCGCCGGCCAAGCCGGCTGTCGTCGCGGCGGTCAAGCCCGTGACGCCGAAGACGGTCGCCCCGGCGCCCGCGCCGGTGAAGGCCGTCCAGCCCGCGCAACCCGCCGCGCCGTTGGCCAAGCCGCCCGTGGTCGCTGCTGCGGCGCCCGCTGCGGCTCCGGCCCCGGCCAAGCCCGCCACCAAGCTTGAACAACTGCAGGCCGCCGTCGCGCCCGCCGCGACCAACGGCGCGGTCCTGGCGACCGGCGAAACCCTCGGTAAGGGCCAGCCTGGCCAAGTGACCCTGTCGCTTCCGGCGACCCTGGGCGAGATGATCCAGAAGGAAGCCGCCAAGCTCGGTTTGGCCAAGGCGGCCAGCAAGACCAGCGCCTATGCCGAACTGCAGGGCGAAGGCTATGAGATCACCCCGAACGGGCGTCAGACGGCGGTCGTGAAGGCGGGCGAGCCGACCACCTTCGCCTGGCAGGTCAAGCCCGGCGCCGAGGCCAAGGGCGAACTGAAGTCGGAGTTCGGTGTCGAACTGACCGGCGCGAAGGAACCGCAAGGCTTCTCCCTGGGTGAGATCAGCAAGCGCGTCGCAGCCCTGCCGCAAGAGGCCAAGAAGGGCCTCGACAAGCTCGACCTCGGCGCCTTGAACGGCACGATCTCGCTGCCCGGCCTTGGCCCGACGCCGATCAAGACCCTGCTGGGTGCGGCTCTGGTGCTTCTGGCGCTGATCATTCTGGTGGGCGTGTCGCGCAGCGTCGCCGCTTCGCGCCGCAAGGCCGAAAGCCAGCGCAAGTACCGCACGCTGACCGACTACGGGCGCAACGAGATGGAGTTCGAGGCGCCTCCGGCCAGCACCCACGTTTCGCACGTGAACCCGTTCCTCGCCGCCGCCGGCGGCGCAGCGGCCGGTGCGGTCGCGGCCTCGGCCGTGGCGCACCATGACGATCACCACGACCACGGGCATGGCCATGATGATCACCACGGCCATGACGACCACGGCCACGGCCACGATGATCATGGCCACGACGCCCACGGTCATGACGATCACGGGCACGGGGCGGATCTTCACGCCCACGCCCACCCTGAACCCCACGCCCCGGCTGAGCACGTTTCGTACGTGAACCCGATGGTGGCCGCGACCACCGCGAGCCACGCCGACCACCATGGGCACGATGCTGCTCACGGGCATGATGATCACGGCCATAGCCACGACGATCATGGTCACGGCCACGACGATCACGGGCATGGTCACGATGATCATGCGCATGGCCACGACGATCATGCCCACGGCCACGACCATGGCGCCCATCACGCCCATGCCGAGCACACCGCCCATGTGACCCCGGCCAGCCATGCCGATGATCACGGCCACGGCCATGACGACCATGGACACGGCCATGACGATCACGGCCATGACGCCCATGGTCACGGCGGCCACGGACATGATGACCACGGCCATCACGAAGACCATGGCCACGGTCACCACGACGCGCCCAAAAAGGAACTTGAGCACGCTCACCAC
PflI
ATGAGCGTCGTCGCCCTGGCCATGAACGGGTTCCTGGCGGTGCTGCTGATCGCGGCGCTGATCTTCGGCTGGCGTCTGGAGCGTCGGCTGAAGGCGCTGCGCGACAGTCACGAAGGCTTCGCCAAGGCGGTCGCCGATCTCGACCAGGCCGCCGCTCGGGCCGAGCAGGGGCTCGCCGATCTCCGCGCCGCCACCGACGAGGCCGCTGAAACCCTGGCCGTCCGGATCGAGCGCGCCCAGGCCCTTGCGGCTCAGCTGGAAGATCGGGTCAACCGGCCTGCGCCGCCCAAGGCTCAGGCAGCGCCCGAACGCGAGGCTCCGCCACGTCCGTCGCGTGCGATCGAGGCTCCGCCGGTCCCGCCCCCTGGCGAGCGTCGCCTTTCGGCGGCCGATTTCGAGCGCCTTCTCGAGCGCGAGGATCGGGTCGAGCGGGCCGCCCGCGGCGAACCGGCGCCGCGACCCGCGCCACGTCCAAATCTGAGCCAAGAAACCCCGCGTTCACGGGCGCGGGTTGATGATGACCTGTTCGATGGGCCGGTTGATCCCCCGCGCCCGACCAGCAATCCACGAGTTCCTCGCCGATGAAGAACATTC
TipF
TTGGGACGGAATGGCGCAGGGACGTTAATCTATTCGACTCTTCAGGCCGGGACCGGTCTGATTGCGTGTAACGATTCGTGTGGACGGTTTATGCGTAGGCTGATGCTGGCGCTCCTGACGGGCGCTTATCTCTGTCTGGCTTTGCTTGTCTCCCTGTTCCTCTTGAGAACGGGAGCGACTCCGTCCGTGGGCGTCTCGGCGTTCATCGGCACGTTGGGCCTGTGCTTTGCGTTCCACGGCCTGATCGCCCAAGCGCTGATGGGCGCGGCCCTGCGCGTCGACATCGACACCATCCGCGAAGCCCACGCCATCCTGCTGGACCAGATCGAGAAGGTCGACGCCCGCGTCACCGACCTCGTCGACACCGTCGCCGCCGACGCCCAGCGCCGCTCCGAGGAGCTGTCCAGCGAAGTCCACCAGCTGGAAGACCTGATCCAGCAGATGAACGACCGCCTGGAGCACCAGCTTACCCACCAGGTCGCCGCCGCCTCGGCCCGCACCGGCGCCCGCGACCGCGCCCCGCAGGCCAGCCACATGCTGCAGGTGGTGCAGGACGCCCTGGCCGAGAACCGGGTCGACCTCTACCTGCAGCCGATCGTCAGCCTGCCCCAGCGCCGCACGGTGTTCTACGAGAGCTTCTCGCGCCTGCGCGATGAGACCGGCCGTGTGATGATGCCGGCCGAGTATCTGGCGGTGGCCGAGCCCGAGGGCCTGATGACGGCCATCGACAACCTCTTGCTGTTCCGCTGCGTGCAGATCGTGCGCCGCCTGGCCAAGCAGGACCGCAAGGTCGGGATCTTCTGCAACATCTCGCTGGCCAGCCTGGGCGACGAGAGCTTCTTCCCGCAGTTCCTGGAGTTCATGCAGGGCAACAAGGACCTGGCCGGGGCGGTGTTCTTCGAGCTGGGCCAGGCGGCGTTCGAGCGTCGCGGTCCGGTCGAGGCCCGCCACATGGCCCGTCTGGCCAGCCTCGGCTTCAGCTTCAGCCTCGACAAGGTCAATGACCTGGACGTCGACTTCCAGGACCTGGCTCGCGCCGACGTGAAGTTCCTGAAAGTCGGCGCCCAGATGATGCTGGACCAGCTGGAAGAGCAGGACGGCAAGCTGGTCATCACCTCGCTGCCCGACCTGAACGCCAGCGACTTCGCCGCCCTCACCCGCCGCTACGGCATCGAGGTGATCGTCGAGAAGGTCGAGGCCGAGAAGCAGGTCGCCGAGGTGCTGGATCTCGACATCGGCTATGGCCAGGGCCACCTGTTCGGCGAGCCGCGCGCCATCCGCGACGCGGTGCTGGCCGAAGCCGATCCGCCCGCCGACTTCATCCGCGCCCCGATGCGTCGCCGCGCCGTGGGCTGG
References
Lab Notebook 8/6/13
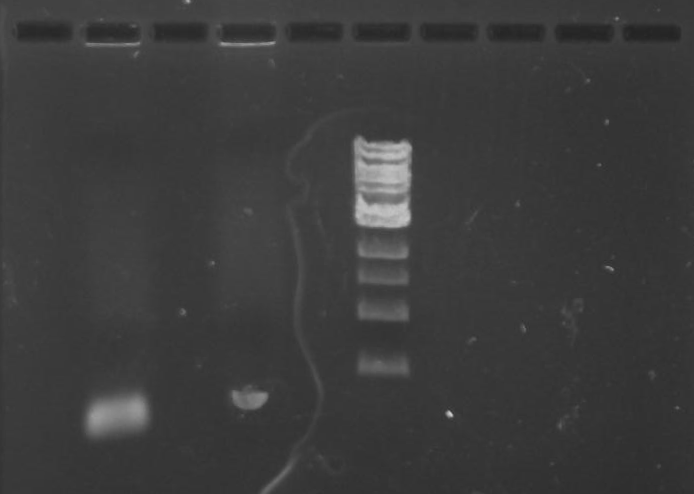
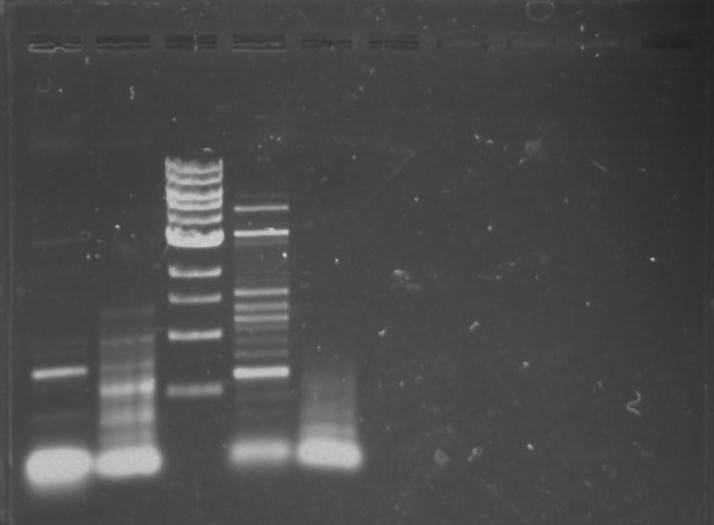
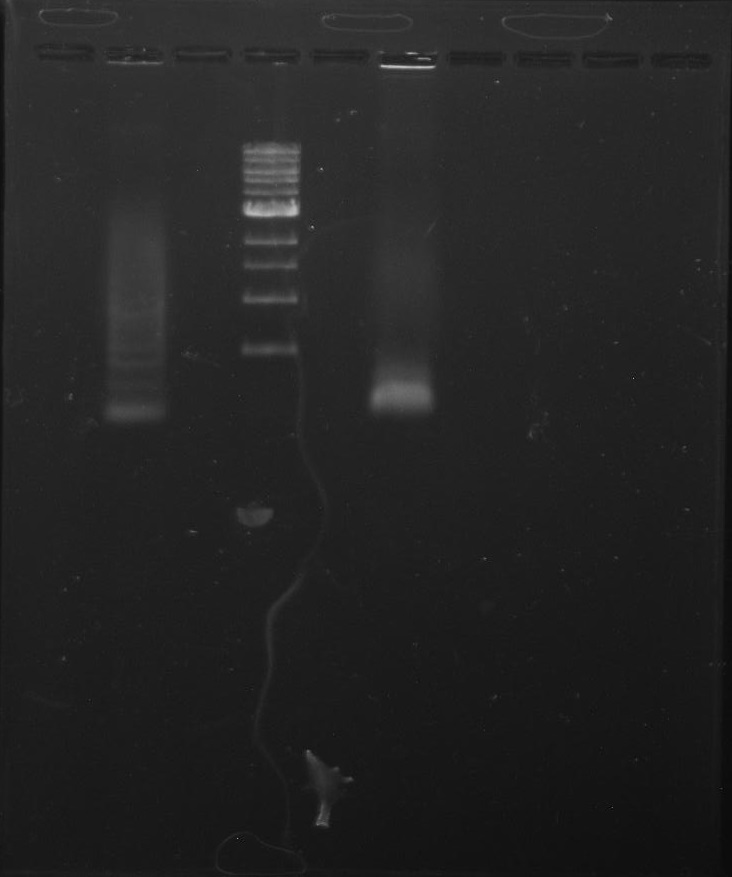
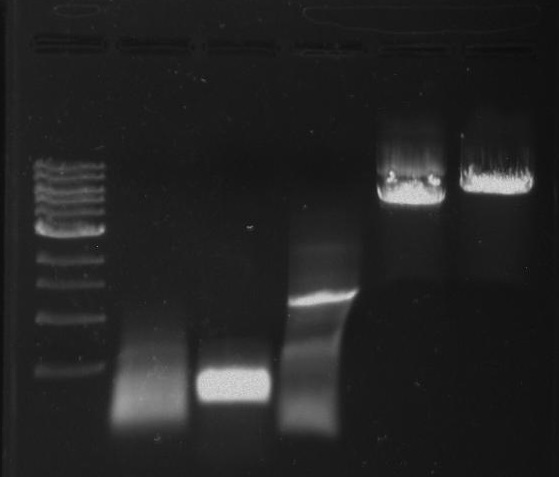
Purpose: To isolate and amplify PflI and Stpx polar markers from Caulobacter Crescentus (CB15N). Expected sizes are 614bp for PflI and 1.9kb for Stpx.
Each PCR sample contains:
- 23 ul MilliQ water
- 1 ul of template (colony pick from CB15 culture in PYE)
- 0.5 ul of 100 uM forward primer Stpx/PflI
- 0.5 ul of 100 uM reverse primer Stpx/PflI
- 25 ul of OneTaq (2x)
Touchdown PCR Protocol:
|
Temp |
94°C |
94°C |
64°C |
68°C |
94°C |
57°C |
68°C |
72°C |
4°C |
|
Time |
3:00 |
0:15 |
0:30 |
2:20 |
0:15 |
0:30 |
2:20 |
5:00 |
Hold |
15 cycles 15 cycles
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 120 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
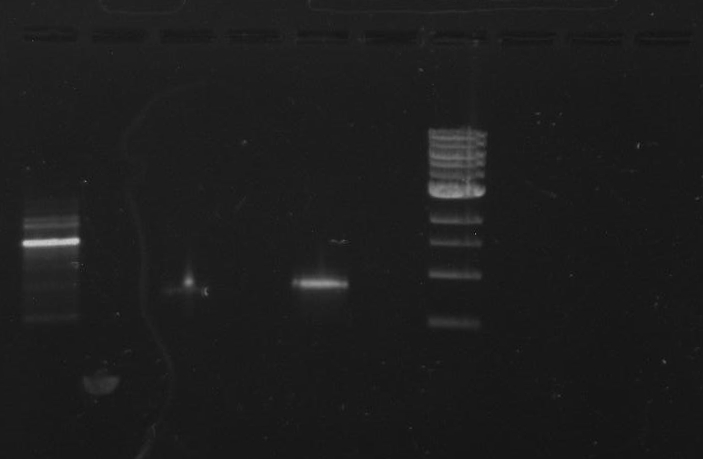
Gel 1: From left to right. PflI, Ladder, Stpx.
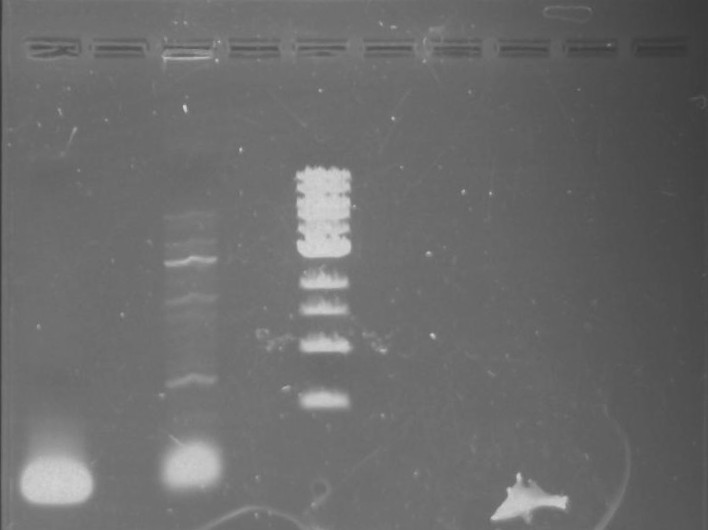
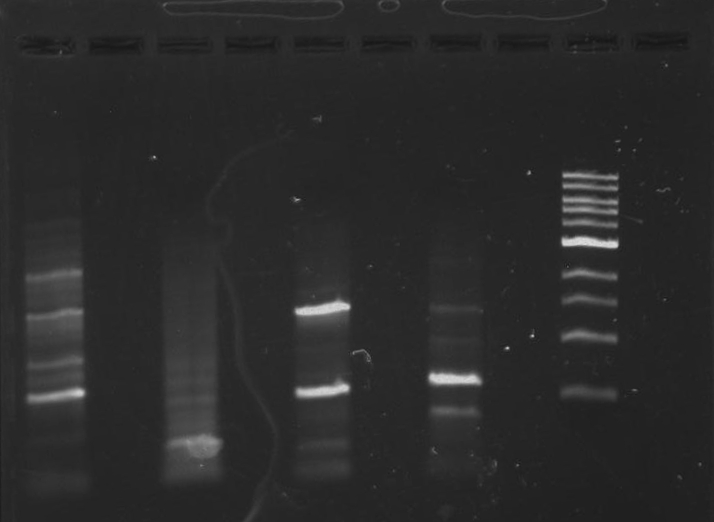
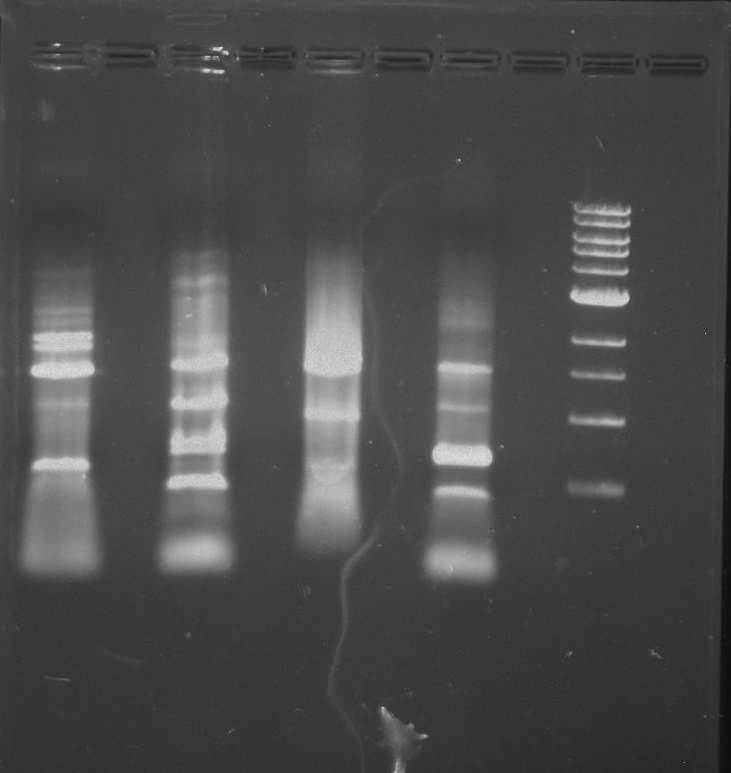
Lab Notebook 8/7/13
Each PCR sample consists:
- 23 ul MilliQ water
- 1 ul of template (colony pick from CB15 culture in PYE)
- 0.5 ul of 100 uM forward primer Stpx/PflI
- 0.5 ul of 100 uM reverse primer Stpx/PflI
- 25 ul of OneTaq (2x)
Touchdown PCR Protocol:
|
Temp |
94°C |
94°C |
60°C |
68°C |
94°C |
53°C |
68°C |
72°C |
4°C |
|
Time |
3:00 |
0:15 |
0:30 |
2:20 |
0:15 |
0:30 |
2:20 |
5:00 |
Hold |
15 cycles 15 cycles
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 120 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Gel 2: From left to right. PflI, Stpx, Ladder.
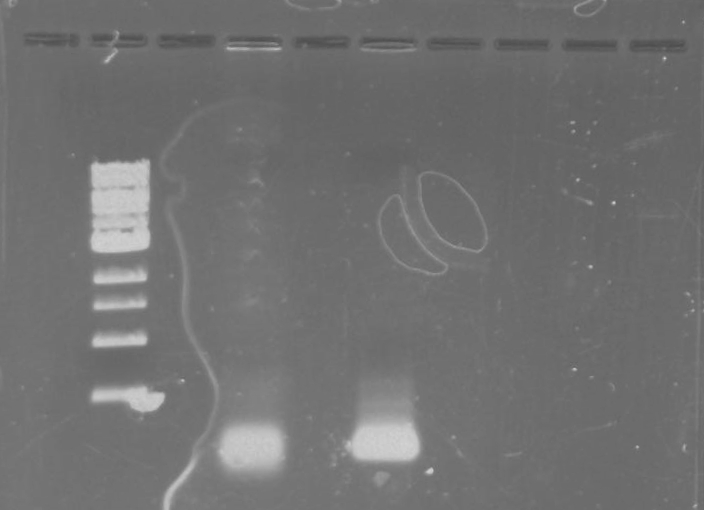
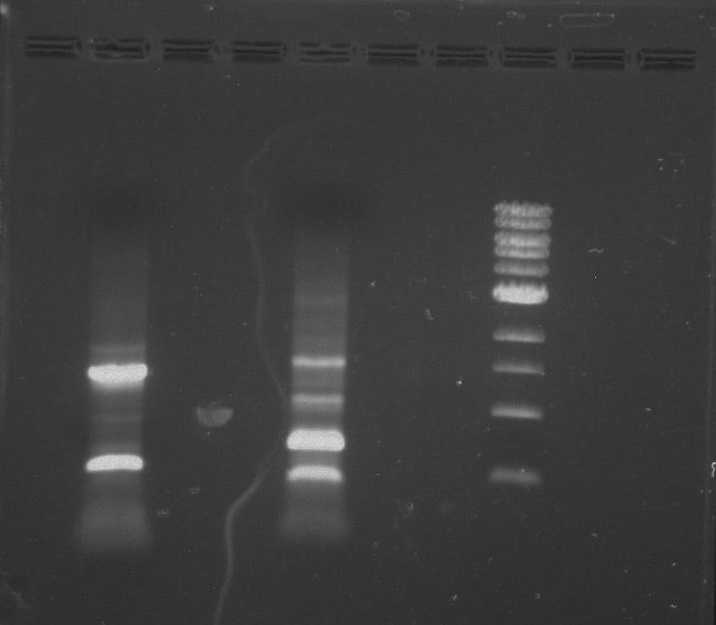
Lab Notebook 8/8/13
Each PCR sample consists:
- 23 ul MilliQ water
- 1 ul of template (colony pick from CB15 culture in PYE)
- 0.5 ul of 100 uM forward primer Stpx/PflI
- 0.5 ul of 100 uM reverse primer Stpx/PflI
- 25 ul of OneTaq (2x)
Touchdown PCR Protocol:
|
Temp |
94°C |
94°C |
56°C |
68°C |
94°C |
49°C |
68°C |
72°C |
4°C |
|
Time |
3:00 |
0:15 |
0:30 |
2:20 |
0:15 |
0:30 |
2:20 |
5:00 |
Hold |
15 cycles 15 cycles
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 120 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Gel 3: From left to right. PflI, Stpx, Ladder.
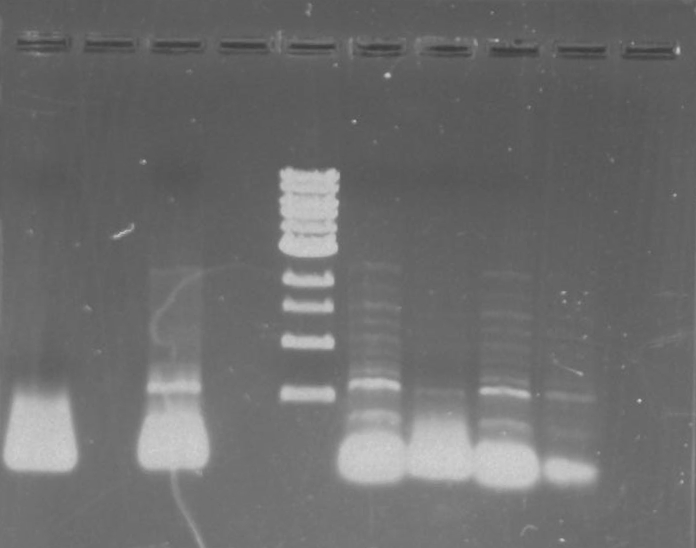
Lab Notebook 8/9/13
Each PCR sample consists:
- 23 ul MilliQ water
- 1 ul of template (colony pick from CB15 culture in PYE)
- 0.5 ul of 100 uM forward primer Stpx/PflI
- 0.5 ul of 100 uM reverse primer Stpx/PflI
- 25 ul of OneTaq (2x)
Touchdown PCR Protocol:
|
Temp |
94°C |
94°C |
56°C |
68°C |
94°C |
49°C |
68°C |
72°C |
4°C |
|
Time |
3:00 |
0:15 |
0:30 |
2:20 |
0:15 |
0:30 |
2:20 |
5:00 |
Hold |
15 cycles 15 cycles
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 120 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Gel 4: From left to right. Ladder, Stpx, PflI.
Lab Notebook 8/13/13
Each PCR sample will contain:
- 9.5 ul MilliQ water
- 1 ul of template (colony pick from CB15N colony pick in PYE)
- 1 ul of forward primer at 100 uM concentration Stpx/PflI
- 1 ul of reverse primer at 100 uM concentration Stpx/PflI
- 12.5 ul of 2x Phusion with GC
PCR Protocol:
|
Temp |
98°C |
98°C |
66°C |
72°C |
72°C |
4°C |
|
Time |
3:00 |
0:10 |
0:15 |
1:20 |
5:00 |
Hold |
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 120 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Gel 5: From left to right. PflI, Stpx, Ladder, PflI, N/A, Stpx, N/A.
Lab Notebook 8/15/13
Two samples of Stpx and PflI were made.
Each PCR sample will contain:
- 23 ul MilliQ water
- 1 ul of template (colony pick from CB15M colony pick in PYE)
- 0.5 ul of PflI/Stpx forward primers at 100 uM concentration
- 0.5 ul of PflI/Stpx reverse primers at 100 uM concentration
- 25 ul of 2x Phusion mix with GC buffer
Touchdown PCR Protocol:
|
Temp |
95°C |
95°C |
56°C |
68°C |
95°C |
49°C |
68°C |
72°C |
4°C |
|
Time |
1:00 |
0:10 |
0:15 |
2:20 |
0:10 |
0:15 |
2:20 |
5:00 |
Hold |
15 cycles 15 cycles
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 120 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Gel 6: From left to right. Stpx, PflI, Ladder, Stpx, PflI.
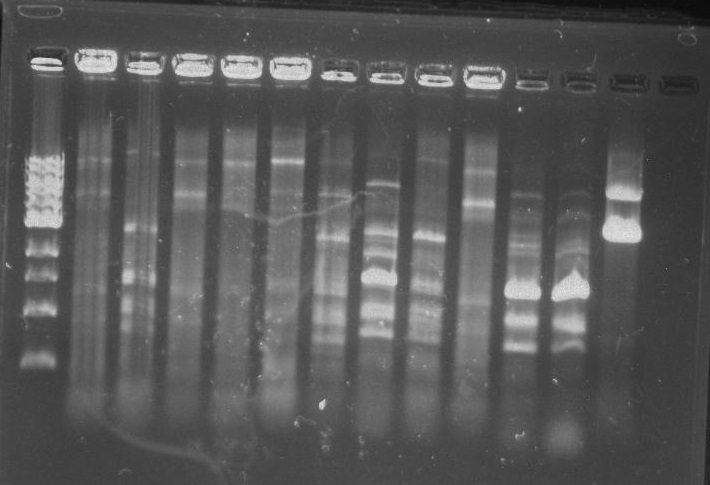
Stpx, PflI, TipF, DivJ PCR:
Four samples will contain:
- 2.5 ul forward primer (Stpx, PflI, DivJ, TipF) at 10 uM concentration
- 2.5 ul reverse primer (Stpx, PflI, DivJ, TipF) at 10 uM concentration
- 1 ul template of CB15N made from a colony pick diluted in water
- 19 ul MilliQ water
- 25 ul of 2x Phusion mix with GC buffer
Touchdown PCR Protocol:
|
Temp |
98°C |
98°C |
56°C |
72°C |
98°C |
49°C |
72°C |
72°C |
4°C |
|
Time |
5:00 |
0:15 |
0:30 |
2:00 |
0:15 |
0:30 |
2:00 |
5:00 |
Hold |
15 cycles 15 cycles
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 120 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Gel 7: from left to right Stpx, PflI, TipF, DivJ, Ladder.
Lab Notebook 8/16/13
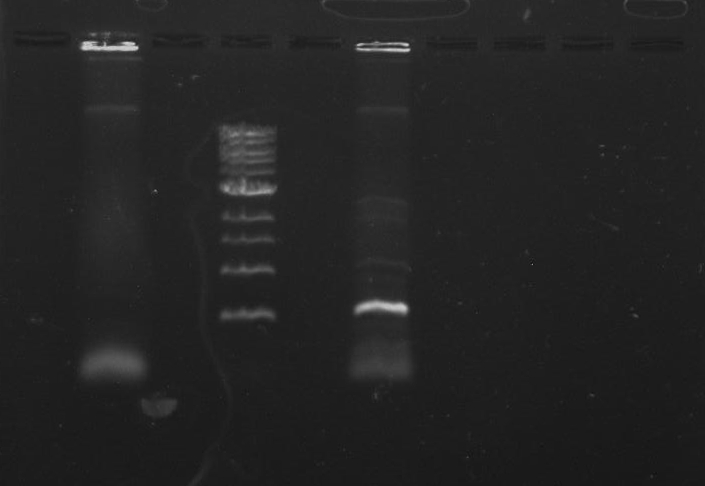
Stpx and TipF PCR:
One Stpx sample and one TipF sample were made.
- 1 ul forward primer (Stpx/TipF) at 10 uM concentration
- 1 ul reverse primer (Stpx/TipF) at 10 uM concentration
- 1 ul template from CB15N glycerol stock
- 9.5 ul MilliQ water
- 12.5 of 2x Phusion mix with GC buffer
Touchdown PCR Protocol:
|
Temp |
98°C |
98°C |
56°C |
72°C |
98°C |
49°C |
72°C |
72°C |
4°C |
|
Time |
5:00 |
0:15 |
0:30 |
2:00 |
0:15 |
0:30 |
2:00 |
5:00 |
Hold |
15 cycles 15 cycles
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 120 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Gel 8: left to right Stpx, ladder, TipF.
Ran the remaining PCR product of TipF and DivJ (45 ul) on a 0.8% agarose gel made from 0.8g of agarose and 100 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 45 ul PCR product + 8 ul 6X gel loading buffer.
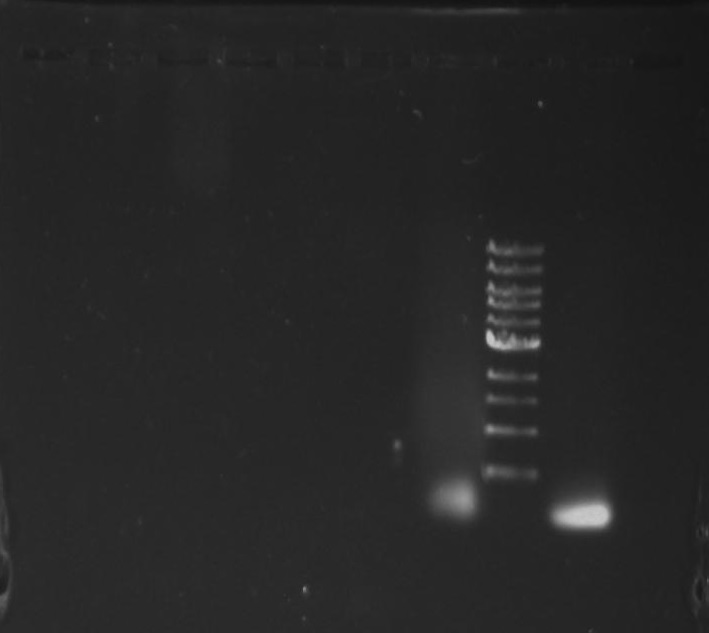
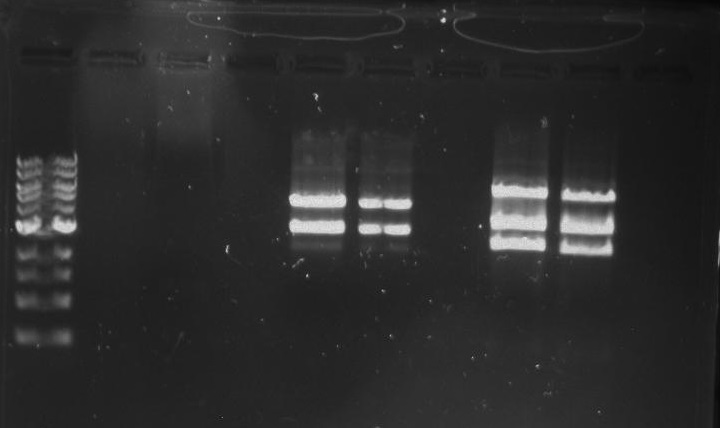
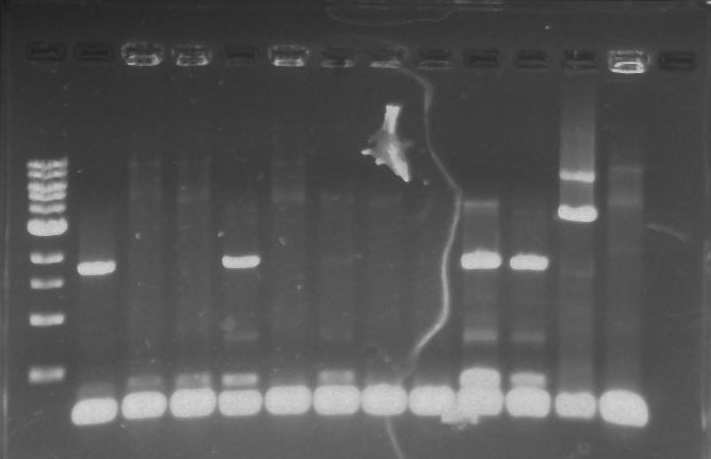
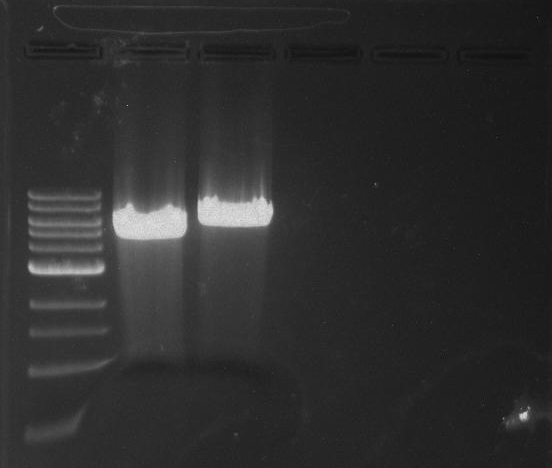
Gel 9: from left to right TipF, DivJ, Ladder.
*gel shows bright bands at expected lengths of ~1.9 kb for TipF and ~600bp for divJ
Purifying DNA from an Agarose Gel:
1) Cut band out in a dark room with a sterile razor and an UV lamp.
2) Place the cut of gel band in a
3) Centrifuge the gel band for 10 minutes at 5,000 xg.
4) Add five volumes of DNA Binding buffer to each volume of DNA sample.
5) Load the mixture into a Zymo-Spin Column in a collection tube.
6) Centrifuge at 10,000 xg for 30 seconds. Discard flow through.
7) Add 200 ul of DNA Wash buffer to the column and centrifuge at 10,000 xg for 30 seconds.
8) Place the Zymo-Spin column into a new 1.5 ml tube.
9) Add 25 ul MilliQ water to column matrix and spin at 10,000 xg for 30 seconds to elute the DNA.
Lab Notebook 8/19/13
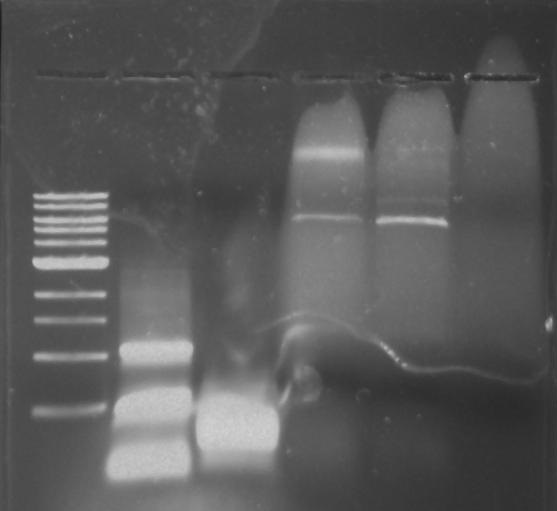
PflI and DivJ PCR:
Two samples:
- 1 ul forward primer (PflI/DivJ) at 10 uM concentration
- 1 ul reverse primer (PflI/DivJ) at 10 uM concentration
- 1 ul template of CB15N made from a colony pick diluted in water
- 9.5 ul MilliQ water
- 12.5 ul of 2x Phusion mix with GC buffer
Touchdown PCR Protocol:
|
Temp |
98°C |
98°C |
56°C |
72°C |
98°C |
49°C |
72°C |
72°C |
4°C |
|
Time |
5:00 |
0:15 |
0:30 |
0:40 |
0:15 |
0:30 |
0:40 |
5:00 |
Hold |
15 cycles 15 cycles
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Gel 10: From left to right. PflI, ladder, DivJ.
Redo TipF and DivJ DNA Purification:
Four PCR samples (x2 TipF and x2 DivJ):
- 2.5 ul forward primer (TipF/DivJ) at 10 uM concentration
- 2.5 ul reverse primer (TipF/DivJ) at 10 uM concentration
- 1 ul template of CB15N glycerol stock
- 19 ul MilliQ water
- 25 ul of 2x Phusion mix with GC buffer
Touchdown PCR Protocol:
|
Temp |
98°C |
98°C |
56°C |
72°C |
98°C |
49°C |
72°C |
72°C |
4°C |
|
Time |
5:00 |
0:15 |
0:30 |
2:00 |
0:15 |
0:30 |
2:00 |
5:00 |
Hold |
15 cycles 15 cycles
Lab Notebook 8/20/13
Gel of 8/19/13 Redo TipF and DivJ:
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.8g of agarose and 100 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 50 ul PCR product + 10 ul 6X gel loading buffer.
Gel 11: From left to right. TipF, DivJ, TipF, DivJ, Ladder.
Purifying DNA from an Agarose Gel:
1) Cut band out in a dark room with a sterile razor and an UV lamp.
2) Place the cut of gel band in a
3) Centrifuge the gel band for 10 minutes at 5,000 xg.
4) Add five volumes of DNA Binding buffer to each volume of DNA sample.
5) Load the mixture into a Zymo-Spin Column in a collection tube.
6) Centrifuge at 10,000 xg for 30 seconds. Discard flow through.
7) Add 200 ul of DNA Wash buffer to the column and centrifuge at 10,000 xg for 30 seconds.
8) Place the Zymo-Spin column into a new 1.5 ml tube.
9) Add 25 ul MilliQ water to column matrix and spin at 10,000 xg for 30 seconds to elute the DNA.
12 PCR Reactions of DivJ and TipF (6 each):
x6 TipF:
- 2.5 ul forward primer TipF at 10 uM concentration
- 2.5 ul reverse primer TipF at 10 uM concentration
- 1 ul template of CB15N glycerol stock
- 19 ul MilliQ water
- 25 ul of 2x Phusion mix with GC buffer
x6 DivJ:
- 2.5 ul forward primer DivJ at 10 uM concentration
- 2.5 ul reverse primer DivJ at 10 uM concentration
- 1 ul template of CB15N glycerol stock
- 19 ul MilliQ water
- 25 ul of 2x Phusion mix with GC buffer
Touchdown PCR Protocol:
|
Temp |
98°C |
98°C |
56°C |
72°C |
98°C |
49°C |
72°C |
72°C |
4°C |
|
Time |
5:00 |
0:15 |
0:30 |
2:00 |
0:15 |
0:30 |
2:00 |
5:00 |
Hold |
15 cycles 15 cycles
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.8g of agarose and 100 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 50 ul PCR product + 10 ul 6X gel loading buffer.
Gel 12: From left to right. TipF, TipF, TipF, TipF, TipF, TipF, Ladder.
Gel 13: From left to right. DivJ, DivJ, DivJ, DivJ, DivJ, DivJ, Ladder.
Lab Notebook 8/21/13
Checking the Purified Samples of TipF and DivJ for One Band:
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Gel 14: From left to right. TipF, DivJ, Ladder.
x2 TipF:
- 2.5 ul forward primer TipF at 10 uM concentration
- 2.5 ul reverse primer TipF at 10 uM concentration
- 1 ul template of CB15N glycerol stock
- 19 ul MilliQ water
- 25 ul of 2x Phusion mix with GC buffer
x2 DivJ:
- 2.5 ul forward primer DivJ at 10 uM concentration
- 2.5 ul reverse primer DivJ at 10 uM concentration
- 1 ul template of CB15N glycerol stock
- 19 ul MilliQ water
- 25 ul of 2x Phusion mix with GC buffer
Touchdown PCR Protocol:
|
Temp |
98°C |
98°C |
56°C |
72°C |
98°C |
49°C |
72°C |
72°C |
4°C |
|
Time |
5:00 |
0:15 |
0:30 |
2:00 |
0:15 |
0:30 |
2:00 |
5:00 |
Hold |
15 cycles 15 cycles
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 50 ul PCR product + 10 ul 6X gel loading buffer.
*Forgot to take a picture of the gel but the gel picture was not good. The TipF samples were not near the expected size. The DivJ samples were smeared.
Lab Notebook 8/22/13
Restriction Digest with NdeI and NsiI
|
DivJ |
TipF |
pXYFPC-2 |
pVCPFPC-4 |
|
28 ul DNA |
35 ul DNA |
5 ul DNA |
5 ul DNA |
|
5 ul 3.1 10x buffer |
5 ul 3.1 10x buffer |
5 ul 3.1 10x buffer |
5 ul 3.1 10x buffer |
|
1 ul NdeI |
1 ul NdeI |
1 ul NdeI |
1 ul NdeI |
|
1 ul NsiI |
1 ul NsiI |
1 ul NsiI |
1 ul NsiI |
|
15 ul MilliQ water |
8 ul MilliQ water |
38 ul MilliQ water |
38 ul MilliQ water |
1) Incubate in the PCR thermocycler at 37°C for 30 minutes then at 65°C for 20 minutes to inactivate.
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.8g of agarose and 100 ml of 1X TBE buffer.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 50 ul PCR product + 10 ul 6X gel loading buffer.
Gel 15: From left to right. Ladder, DivJ with NotI, DivJ without NotI, pXYFPC-2, pVCPFPC-4.
Purifying Vector DNA from an agarose gel
1) Cut band out in a dark room with a sterile razor and an UV lamp.
2) Place the cut of gel band in an agarose spin column.
3) Centrifuge the gel band for 10 minutes at 5,000 xg.
4) Add five volumes of DNA Binding buffer to each volume of DNA sample.
5) Load the mixture into a Zymo-Spin Column in a collection tube.
6) Centrifuge at 10,000 xg for 30 seconds. Discard flow through.
7) Add 200 ul of DNA Wash buffer to the column and centrifuge at 10,000 xg for 30 seconds.
8) Place the Zymo-Spin column into a new 1.5 ml tube.
9) Add 25 ul MilliQ water to column matrix and spin at 10,000 xg for 30 seconds to elute the DNA.
NotI and DivJ Restriction Digest:
- 12.5 ul DivJ DNA
- 5 ul CutSmart 10X buffer
- 1 ul NotI
- 31.5ul MilliQ water
1) Incubate in the PCR thermocycler at 37°C for 30 minutes.
Refer to Gel 15.
Lab Notebook 8/23/13
DivJ and TipF Ligations
|
Mastermix for pXYFPC-2 |
Mastermix for pVCPFPC-4 |
|
12 ul of pXYFPC-2 |
12 ul of pVCPFPC-4 |
|
3 ul of 10x T4 buffer |
3 ul of 10x T4 buffer |
|
1.5 ul T4 Ligase |
1.5 ul T4 Ligase |
Six total ligation reactions:
|
1) 5.5 ul of Mastermix for pXYFPC-2 + 4.5 ul of DivJ |
4) 5.5 ul of Mastermix for pVCPFPC-4 + 4.5 ul of DivJ |
|
2) 5.5 ul of Mastermix for pXYFPC-2 + 4.5 ul of MilliQ water |
5) 5.5 ul of Mastermix for pVCPFPC-4 + 4.5 ul of MilliQ water |
|
3) 5.5 ul of Mastermix for pXYFPC-2 + 4.5 ul of TipF |
6) 5.5 ul of Mastermix for pVCPFPC-4 + 4.5 ul of TipF |
Incubate ligation reactions for 30 minutes or greater at room temperature.
Heat Shock Transformations:
1) Add 40 ul of Top10 Chemicompetent cells in 10 ul of each ligation reaction.
2) Place in a 42°C water bath for 30 seconds.
3) Place back on ice for 5 minutes.
4) Add 200 ul of LB to a microcentrifuge tube.
5) Transfer the Top10 Chemicompetent cells and ligation reaction to the microcentrifuge tube.
6) Incubate for one hour at 37°C at 200 rpm.
Plating the Transformations:
1) Obtain three plates of LB and gentamicin and three plates of LB and kanamycin.
2) Warm up the plates in the 37°C incubator prior to plating.
3) Pipet all of the transformation into the plate.
4) Using a bunsen burner, heat up the steel rod.
5) Create streaks with the steel rod.
6) Wait for 5 minutes until all the liquid is soaked up by the plate.
7) Tape up the plates and/or place the plates in a plastic bag.
8) Place plates in an incubator at 37°C.
Lab Notebook 8/25/13
Created an overnight of the DivJ and TipF plates by picking five colonies from each LB and gentamicin plate plus one colony from the negative control. The colony picks were diluted in 20 ul of MilliQ water.
Lab Notebook 8/26/13
Purpose: colony plasmid isolation
Miniprep of TipF and DIvJ:
1) Add 100 ul of 7X Lysis buffer to 600 ul of e. coli culture in a 1.5 ml microcentrifuge tube. Mix by inverting the tube 4-6 times and lyse samples for 1-2 minutes.
2) Add 350 ul of cold Neutralization buffer. Mix thoroughly. Neutralization is complete when sample becomes yellow and precipitate has formed.
3) Centrifuge at 16,000 xg for 2 minutes.
4) Transfer the supernatant into the Zymo-Spin IIN column .
5) Place column into a collection tube and centrifuge at 11,000 xg for 15 seconds. Discard the flow through and place the column back into the same collection tube.
6) Add 200 ul of ENdo Wash buffer to the column. Centrifuge at 11,000 xg for 30 seconds.
7) Add 400 ul of Zyppy Wash buffer to the column. Centrifuge at 11,000 xg for 30 seconds.
8) Transfer the column into a clean 1.5 ml microcentrifuge tube then add 30 ul of MilliQ water. Let stand for one minute at room temperature. Centrifuge at 11,000 xg for 15 seconds to elute the DNA.
Purpose: diagnostic PCR reactions to amplify target sequences
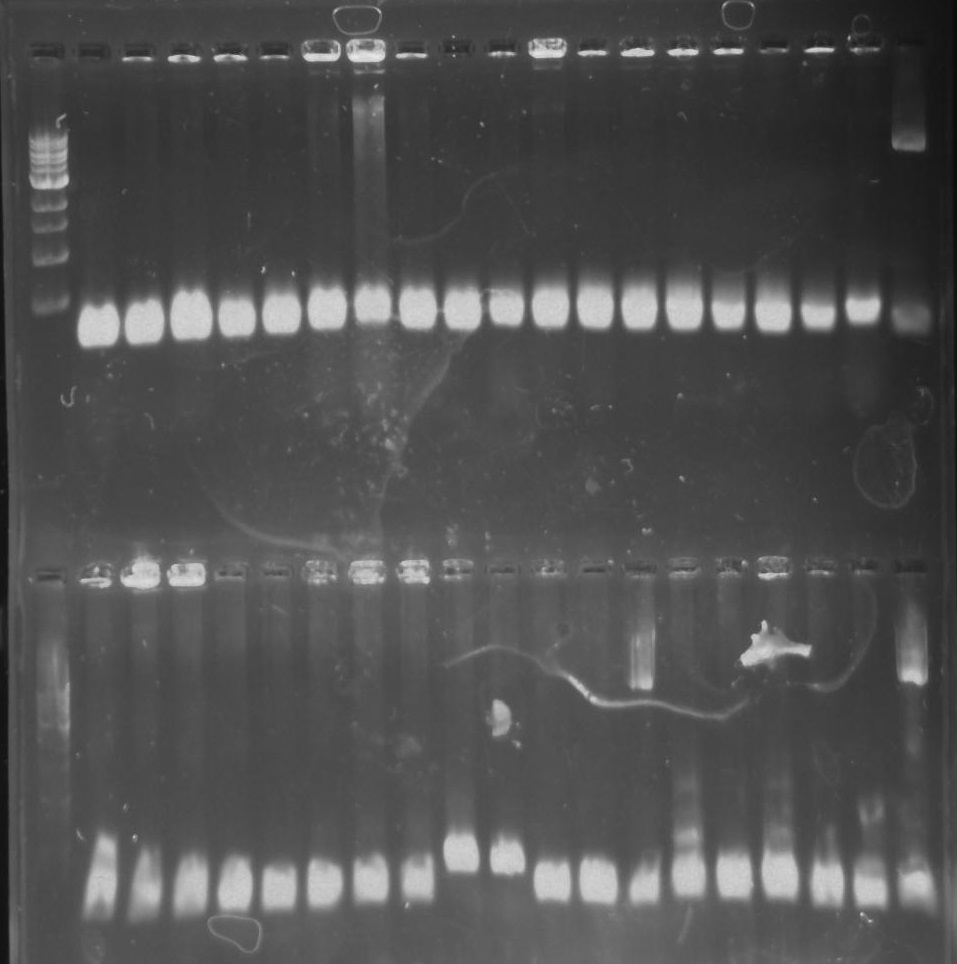
PCR:
x5 TipF
- 1 ul of eluted TipF DNA
- 1 ul of forward primer TipF at 10 uM concentration
- 1 ul of reverse primer TipF at 10 uM concentration
- 2 ul MilliQ water
- 5 ul of 2x phusion mix with GC buffer
x5 DivJ
- 1 ul of eluted DivJ DNA
- 1 ul of forward primer DivJ at 10 uM concentration
- 1 ul of reverse primer DivJ at 10 uM concentration
- 2 ul MilliQ water
- 5 ul of 2x phusion mix with GC buffer
x2 Positive Control
- 1 ul of CB15N glycerol stock
- 1 ul of forward primer DivJ/TipF at 10 uM concentration
- 1 ul of reverse primer DivJ/TipF at 10 uM concentration
- 2 ul MilliQ water
- 5 ul of 2x phusion mix with GC buffer
x2 Negative Control
- 1 ul of colony pick from the negative control plate
- 1 ul of forward primer DivJ/TipF at 10 uM concentration
- 1 ul of reverse primer DivJ/TipF at 10 uM concentration
- 2 ul MilliQ water
- 5 ul of 2x phusion mix with GC buffer
Touchdown PCR Protocol:
|
Temp |
98°C |
98°C |
56°C |
72°C |
98°C |
49°C |
72°C |
72°C |
4°C |
|
Time |
5:00 |
0:15 |
0:30 |
2:00 |
0:15 |
0:30 |
2:00 |
5:00 |
Hold |
15 cycles 15 cycles
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.46g of agarose and 60 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 10 ul PCR product + 2 ul 6X gel loading buffer.
Gel 16: From left to right: Ladder, TipF1, TipF2, TipF3, TipF4, TipF5, Negative Control from TipF plate, Positive Control, DivJ1, DivJ2, DivJ3, DivJ4, DivJ5, Negative Control from DivJ plate, Positive. Control.
Sequencing:
We sent in TipF5 and DivJ3 and DivJ4 in for sequencing.
For TipF5, we used 10 ul of the miniprepped TipF DNA and 3 ul of the forward primer at 1 uM concentration. Also 10 ul of the miniprepped TipF DNA and 3 ul of the reverse primer at 1 uM concentration.
For DivJ3, we used 10 ul of the miniprepped DivJ DNA and 3 ul of the forward primer at 1 uM concentration. Also 10 ul of the miniprepped DivJ DNA and 3 ul of the reverse primer at 1 uM concentration.
For DivJ4, we used 10 ul of the miniprepped DivJ DNA and 3 ul of the forward primer at 1 uM concentration. Also 10 ul of the miniprepped DivJ DNA and 3 ul of the reverse primer at 1 uM concentration.
Lab Notebook 8/27/13
x10 TipF samples:
- 1 ul of eluted miniprepped TipF DNA
- 1 ul of forward primer TipF at 10 uM concentration
- 1 ul of reverse primer TipF at 10 uM concentration
- 2 ul MilliQ water
- 5 ul of 2x phusion mix with GC buffer
x10 DivJ samples:
- 1 ul of eluted miniprepped DivJ DNA
- 1 ul of forward primer DivJ at 10 uM concentration
- 1 ul of reverse primer DivJ at 10 uM concentration
- 2 ul MilliQ water
- 5 ul of 2x phusion mix with GC buffer
Touchdown PCR Protocol:
|
Temp |
98°C |
98°C |
56°C |
72°C |
98°C |
49°C |
72°C |
72°C |
4°C |
|
Time |
5:00 |
0:15 |
0:30 |
2:00 |
0:15 |
0:30 |
2:00 |
5:00 |
Hold |
15 cycles 15 cycles
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.8g of agarose and 100 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 10 ul PCR product + 2 ul 6X gel loading buffer.
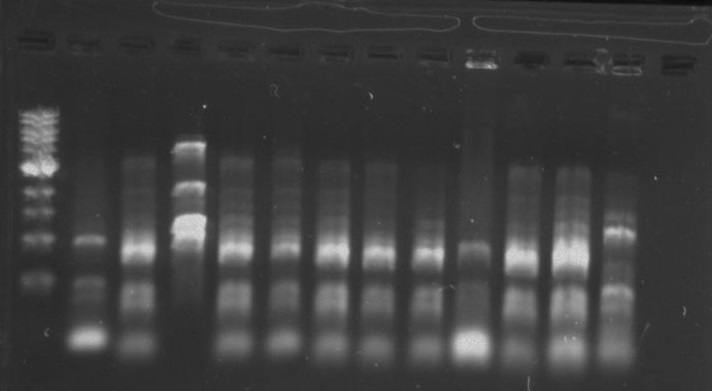
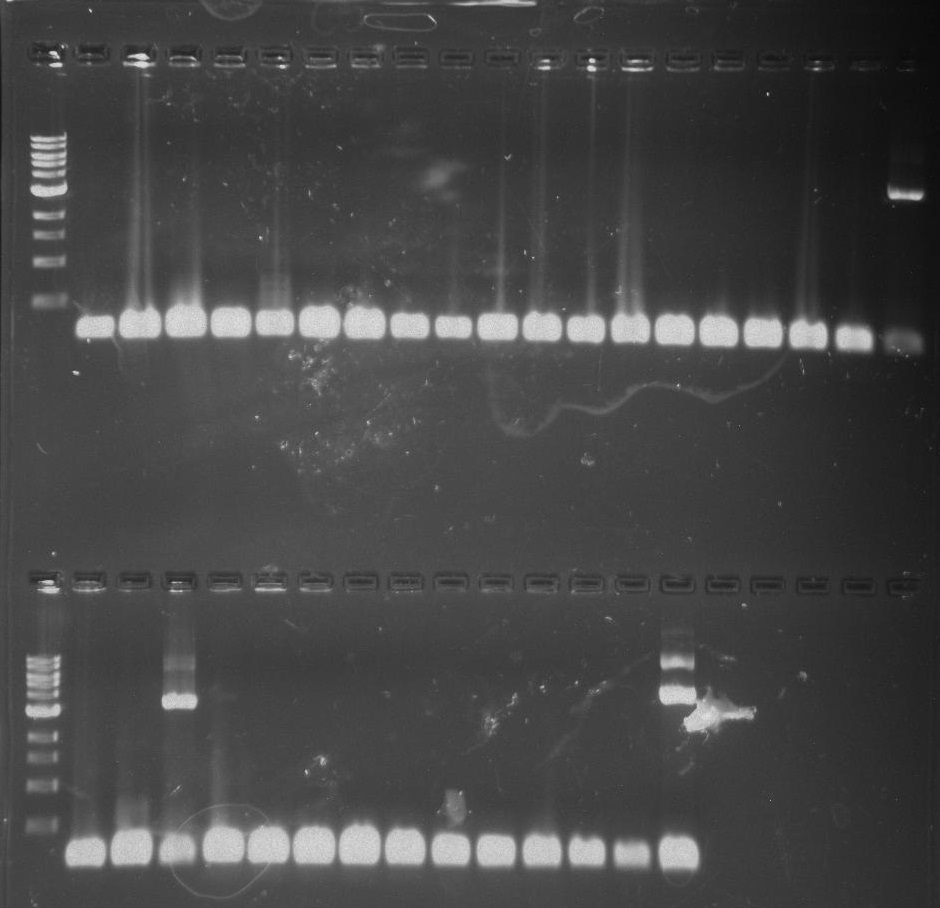
Gel 17: From left to right. Ladder, TipF1, TipF2, TipF3, TipF4, TipF5,TipF6, TipF7, TipF8, TipF9, TipF10, Negative Control from TipF plate, Positive Control.
*TipF3 shows a band at expected length
Gel 18: From left to right. Ladder, DivJ1, DivJ2, DivJ3, DivJ4, DivJ5, DivJ6, DivJ7, DivJ8, DivJ9, DivJ10, Negative Control from DivJ plate, Positive. Control.
Overnight culture of TipF3 for Miniprep:
From the colony pick from the LB + gentamicin TipF plate, we diluted it in 20 ul of MilliQ water. That mixture was then transferred into 4 ml of LB. 6 ul of gentamicin antibiotic was added. The overnight culture was stored in the incubator at 28°C at 220 rpm.
Lab Notebook 8/28/13
Miniprep of Overnight TipF3 Culture:
Spin down overnight culture for 10 minutes at 3,000 xg. Discard 3.4 ml of the supernatant. Resuspend the pellet.
1) Add 100 ul of 7X Lysis buffer to 600 ul of e. coli culture in a 1.5 ml microcentrifuge tube. Mix by inverting the tube 4-6 times and lyse samples for 1-2 minutes.
2) Add 350 ul of cold Neutralization buffer. Mix thoroughly. Neutralization is complete when sample becomes yellow and precipitate has formed.
3) Centrifuge at 16,000 xg for 2 minutes.
4) Transfer the supernatant into the Zymo-Spin IIN column .
5) Place column into a collection tube and centrifuge at 11,000 xg for 15 seconds. Discard the flow through and place the column back into the same collection tube.
6) Add 200 ul of ENdo Wash buffer to the column. Centrifuge at 11,000 xg for 30 seconds.
7) Add 400 ul of Zyppy Wash buffer to the column. Centrifuge at 11,000 xg for 30 seconds.
8) Transfer the column into a clean 1.5 ml microcentrifuge tube then add 30 ul of MilliQ water. Let stand for one minute at room temperature. Centrifuge at 11,000 xg for 15 seconds to elute the DNA.
Sequencing of TipF3:
- 10 ul of miniprepped TipF + 3 ul of TipF forward primer at 1 uM concentration
- 10 ul of miniprepped TipF + 3 ul of TipF reverse primer at 1 uM concentration
Lab Notebook 8/29/13
pVan-for and eGYC-1 PCR of TipF Colony Picks:
Mastermix:
- 60 ul of 2x Phusion mix with GC buffer
- 24 ul MilliQ water
- 12 ul pVan-for primer at 10 uM concentration
- 12 ul eGYC-1 primer at 10 uM concentration
Colony Picks:
- Colony pick of TipF in the LB and gentamicin plate into 20 ul of MilliQ water.
Twelve PCR Reactions:
|
TipF 1-10 |
Negative Control |
Positive Control |
|
9 ul of Mastermix |
9 ul of Mastermix |
9 ul of Mastermix |
|
1 ul of colony pick in water |
1 ul of Colony pick in water |
1 ul of pYCFPC-4 |
Touchdown PCR Protocol:
|
Temp |
98°C |
98°C |
60°C |
72°C |
98°C |
53°C |
72°C |
72°C |
4°C |
|
Time |
5:00 |
0:15 |
0:30 |
2:00 |
0:15 |
0:30 |
2:00 |
5:00 |
Hold |
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.46g of agarose and 60 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 10 ul PCR product + 2 ul 6X gel loading buffer.
Gel 19: From left to right: Ladder, Gel Protocol: We created an 0.8% agarose gel by obtaining 0.4g of agarose and 50 ml of 1X TBE buffer.
Lab Notebook 8/30/13
x2 DivJ samples:
- 2.5 ul forward primer DivJ at 10 uM concentration
- 2.5 ul reverse primer DivJ at 10 uM concentration
- 1 ul template of CB15N glycerol stock
- 19 ul MilliQ water
- 25 ul of 2x Phusion mix with GC buffer
Negative Control:
- 20 ul MilliQ water
- 2.5 ul forward primer DivJ at 10 uM concentration
- 2.5 ul reverse primer DivJ at 10 uM concentration
- 25 ul of 2x Phusion mix with GC buffer
Touchdown PCR Protocol:
|
Temp |
98°C |
98°C |
64°C |
72°C |
98°C |
57°C |
72°C |
72°C |
4°C |
|
Time |
5:00 |
0:15 |
0:30 |
1:00 |
0:15 |
0:30 |
1:00 |
5:00 |
Hold |
15 cycles 15 cycles
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.8g of agarose and 100 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 50 ul PCR product + 10 ul 6X gel loading buffer.
Gel 20: From left to right. Ladder, DivJ1, Remaining DivJ1, DivJ2.
Lab Notebook 8/31/13
Miniprep of TipF.
Lab Notebook 9/1/13
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.4g of agarose and 50 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 50 ul PCR product + 10 ul 6X gel loading buffer.
Gel 21: From left to right. TipF, TipF Ladder.
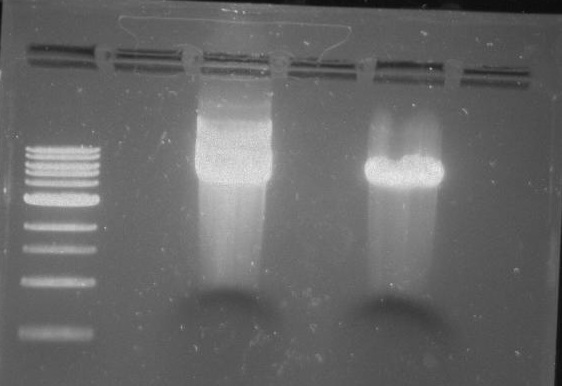
Gel 22: From left to right. Ladder, Vector C, Vector Y.
Cut out the vectors and purify along with DivJ.
Ligation:
|
Mastermix for pXYFPC-2 |
Mastermix for pVCPFPC-4 |
|
7 ul Vector C |
7 ul Vector Y |
|
2 ul of 10x T4 buffer |
2 ul of 10x T4 buffer |
|
1 ul of T4 Ligase |
1 ul of T4 Ligase |
Four total ligation reactions:
|
1) pXYFPC-2 Mastermix + 5 ul MilliQ water |
3) pVCPFPC-4 + 5 ul MilliQ water |
|
2) pXYFPC-2 Mastermix + 326 DivJ |
4) pVCPFPC-4 + 326 DivJ |
Heat Shock Transformations:
1) Add 100 ul of BL21 Chemicompetent cells in 10 ul of each ligation reaction.
2) Place in a 42°C water bath for 30 seconds.
3) Place back on ice for 5 minutes.
4) Add 200 ul of LB to a microcentrifuge tube.
5) Transfer the BL21 Chemicompetent cells and ligation reaction to the microcentrifuge tube.
6) Incubate for one hour at 37°C at 200 rpm.
Plating the Transformations:
1) Obtain two plates of LB and gentamicin and three plates of LB and kanamycin.
2) Warm up the plates in the 37°C incubator prior to plating.
3) Pipet all of the transformation into the plate.
4) Add glass beads to the plates and shake.
5) Remove the glass beads and incubate overnight at 37°C.
Lab Notebook 9/2/13
PCR of the 326 DivJ:
Same protocol as 8/30/13
Purified 326 DivJ by cutting it out of the gel and doing a clean and concentrate.
Lab Notebook 9/3/13
Transformation #3
Purpose: To insert DivJ into Vectors and transform ligated fusion vectors/DivJ into competent cells.
Work Flow: Restrict plasmids Y and C. Run restricted plasmids and DivJ(PCR product) on agarose gel. Cut out DNA/ Clean DNA/ Concentrate DNA. Restriction digest of DivJ. Clean and Concentrate DivJ. Ligate sequences. Transform into electrocompetent cells (e.coli top ten).
Restrict plasmids
Vector C
5 uL DNA
5 uL CutSmart (10x) buffer
1 uL NdeI
1 uL AclI
38 uL MilliQ water
Vector Y
5 uL DNA
5 uL CutSmart (10x) buffer
1 uL NdeI
1 uL AclI
38 uL MilliQ water
-incubate at 37° C for 30 min.
Agarose Gel
Gel 23: From left to right. 326 DivJ, Vector Y, Vector C.
-cut bands out of gel
-clean/concentrate
-elute vectors to 12 ul
-elute DivJ to 30 ul
Restriction Digest (DivJ)
30 ul DivJ DNA
5 ul CutSmart (10x) buffer
1 ul NdeI
1 ul AclI
13 ul MilliQ water
-incubate at 37° C for 30 min.
-clean/concentrate
-elute to 10 ul
Ligation reactions
Vec C master mix (MMC) Vec Y master mix (MMY)
7 ul vector C 7 ul vector Y
2 ul T4 buffer 2 ul T4 buffer
1 ul T4 ligase 1 ul T4 ligase
-prepare reactions
Neg Control C (NC) Vec C w/ DivJ insert (CD)
5 ul MMC 5 ul MMC
5ul MilliQ H2O 5 ul DivJ
Neg Control Y (NY) Vec Y w/ DivJ insert (NC)
5 ul MMY 5 ul MMY
5 ul MilliQ H2O 5 ul DivJ
- incubate at room temp for 30 min.
Electroporate
Top10 electrocompetent cells
-for each ligation product
250 ul top 10 electrocompetent cells
10 ul ligation reaction
*note too much salt
-recovery in 1 ml LB @ 37° C 1 hour
-plate on LB/kan plates for Y vectors and LB/gen plates for C vectors
Lab Notebook 9/4-5/13
Synthesized the minimal DivJ by:
Creating an oligo mix consisting of 5 ul of each oligo at 100 uM concentration.
Synthesize PCR protocol:
- 1 ul of oligo mix
- 0.5 ul forward primer at 100 uM concentration (NdeI)
- 0.5 ul reverse primer at 100 uM concentration (AciI)
- 23 ul MilliQ water
- 25 ul 2X phusion mix with GC buffer
|
Temp |
98°C |
98°C |
63°C |
72°C |
72°C |
4°C |
|
Time |
0:30 |
0:10 |
0:10 |
0:30 |
5:00 |
Hold |
Amplify PCR Protocol:
- 1 ul of the synthesis PCR product
- 0.5 ul forward primer at 100 uM concentration (NdeI)
- 0.5 ul reverse primer at 100 uM concentration (AciI)
- 23 ul MilliQ water
- 25 ul 2X phusion mix with GC buffer
|
Temp |
98°C |
98°C |
63°C |
72°C |
72°C |
4°C |
|
Time |
0:30 |
0:10 |
0:10 |
0:30 |
5:00 |
Hold |
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.4g of agarose and 50 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 50 ul PCR product + 10 ul 6X gel loading buffer.
Gel 24: From left to right. Ladder, Synth PCR, Amp PCR, DivJ, Vector C digested, Vector Y digested.
Lab Notebook 9/6/13
Electroporation but this time with BL21 electrocompetent cells.
Plates from this did not look good as well.
* note BL21 was not a good choice for transformations due to the presence of RecA gene.
Lab Notebook 9/7/13
Purpose: prepare top 10 chemi competent cells for use in future transformations.
* note bad results from transformation attempts with top ten electrocompetent cells may have been due to salty ligations and low yields. The chemi competent cells may allow us to use more of our ligation products during transformations.
Making Top10 Chemicompetent Cells:
1) Grow Top10 SOB overnight between the temperatures 20°C and 33°C.
2) Measure the OD and bring the OD back to 0.1.
3) Add 2.5 ml of Top10 SOB to SOB.
4) Incubate at 31°C for one hour and 20 minutes or until the OD is 0.6.
5) Create 5 ml of 1X Washing buffer: 2.5 ml of Dilution buffer and 2.5 ml of Wash buffer (2X)
6) Create 5 ml of 1X Competent buffer: 2.5 ml Dilution buffer and 2.5 ml of Competent buffer (2X)
7) Incubate the culture on ice for ten minutes and then pellet cells at 2,500 xg for ten minutes at 0-4°C.
8) Remove supernatant. Gently resuspend cells in 5 ml of ice cold 1X Wash buffer. Re-pellet as in step 7.
9) Remove supernatant completely. Gently resuspend cells in 5 ml of ice cold 1X Competent buffer.
10) Aliquot 100-200 ul. Flash freeze.
11) Store in -80°C freezer.
Making LB+Kanamycin and LB+Gentamicin Agar Plates:
To make a 500 ml batch:
- 475 ml of MilliQ water
- 5 g of Tryptone
- 5 g of NaCl
- 2.5 g of yeast Extract
- 7.5 g of Agar
We made x2 500 ml batches.
Combine the reagents and shake until the solutes have dissolved. Adjust the ph to 7.0. Sterilize by autoclaving for 20 minutes on liquid cycle.
Take out of the autoclave and let the solution cool. Place in a water bath to prevent solid formation.
Add 1.25 ml of gentamicin antibiotic to one of the 500 ml batches and 500 ul of kanamycin to the other batch.
Pour into petri dishes about half way. Let agar solidify. Place in the refrigerator.
Lab Notebook 9/8/13
Testing the Top10 Chemicompetent Cells:
We tested our Top10 Chemicompetent cells by using different amount of DNA. We used 300 ng of DNA and 30 ng of DNA from our vec C and Vec Y plasmid stocks. The 300 ng of DNA plates had more growth than the 30 ng of DNA plates.
Transformations
K1 G1
1 ul dilute vector Y 1 ul dilute vector C
100 ul CC top 10 cells 100 ul CC top 10 cells
K2 G2
1 ul Vector Y 1 ul Vector C
100 ul CC top 10 cells 100 ul CC top 10 cells
KN GN
1 ul MilliQ H2O 1 ul MilliQ H2O
100 ul CC top 10 cells 100 ul CC top 10 cells
Lab Notebook 9/9/13
Top10 Chemi competent Cell Transformation of DivJ
Heat Shock Transformations:
1) Add 100 ul of Top10 Chemicompetent cells in 10 ul of each ligation reaction.
2) Place in a 42°C water bath for 30 seconds.
3) Place back on ice for 5 minutes.
4) Add 500 ul of SOC to a microcentrifuge tube.
5) Transfer the Top10 Chemicompetent cells and ligation reaction to the microcentrifuge tube.
6) Incubate for one hour at 37°C at 200 rpm.
Colony PCR of TipF
Mastermix:
- 60 ul of 2x OneTaq
- 24 ul MilliQ water
- 12 ul new-pVan-for primer at 10 uM concentration
- 12 ul new-eGYC-1 primer at 10 uM concentration
Colony Picks:
- Colony pick of TipF in the LB and gentamicin plate into 20 ul of MilliQ water.
Twelve PCR Reactions:
|
TipF 1-10 |
Negative Control |
Positive Control |
|
9 ul of Mastermix |
9 ul of Mastermix |
9 ul of Mastermix |
|
1 ul of colony pick in water |
1 ul of Colony pick in water |
1 ul of pYCFPC-4 |
Touchdown PCR Protocol:
|
Temp |
94°C |
94°C |
60°C |
68°C |
94°C |
53°C |
68°C |
68°C |
4°C |
|
Time |
5:00 |
0:15 |
0:30 |
2:00 |
0:15 |
0:30 |
2:00 |
5:00 |
Hold |
15 cycles 15 cycles
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 10 ul PCR product + 2 ul 6X gel loading buffer.
Gel 25: From left to right. Ladder, TipF1, TipF2, TipF3, TipF4, TipF5,TipF6, TipF7, TipF8, TipF9, TipF10, Negative Control from TipF plate, Positive Control.
Lab Notebook 9/10/13
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.8g of agarose and 100 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 10 ul PCR product + 2 ul 6X gel loading buffer.
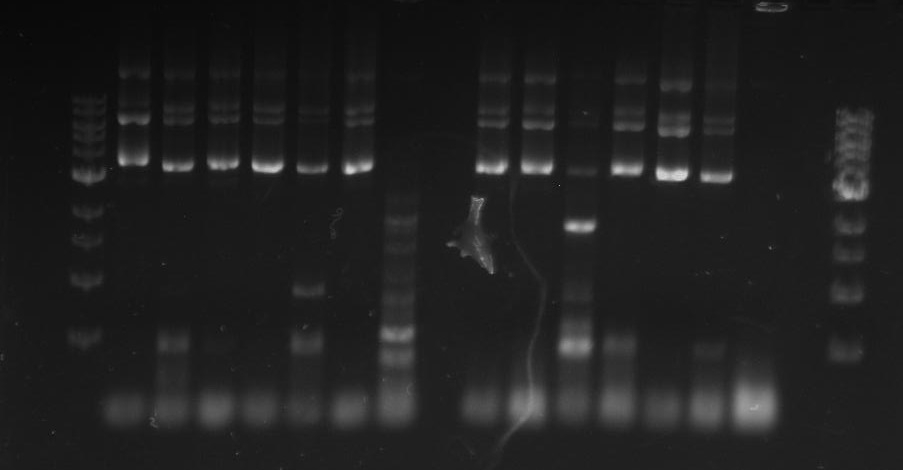
Gel 26: From left to right. Top: Ladder, T1, T2, T3, T4, T5, T6, T7, Y8, T9, T10, CD1, CD2, CD3, CD4, CD5, CD6, CD7, NC, PC. Botton: Ladder, CD8, CA1, CA2, CA3, CA4, CA5, CA6, CA7, CA8, CA9, CA10, NC, PC, YD1, YD2, YD3, YD4, NY, PY. T = TipF, CD = 326 DivJ in pVCPFPC-4. CA = Minimal DivJ in pVCPFPC-4. YD = 326 DivJ in pXYFPC-2. NC = Negative control in pVCPFPC-4, PC = Positive control in pVCPFPC-4, NY = negative control in pXYFPC-2, PY = positive control in pXYFPC-2.
Lab Notebook 9/11/13
PCR of 326 DivJ and Minimal DivJ:
326 DivJ:
1 ul template from CB15N glycerol stock
2.5 ul DivJ forward primer at 10 uM concentration (NdeI)
2.5 ul DivJ reverse primer at 10 uM concentration (AciI)
19 ul MilliQ water
25 ul of 2x phusion mix with GC buffer
Minimal DivJ:
1 ul of Synthesis PCR product
0.5 ul DivJ forward primer at 100 uM concentration (248)
0.5 ul DivJ reverse primer at 100 uM concentration (328)
23 ul MilliQ water
25 ul of 2x phusion mix with GC buffer
Touchdown PCR Protocol:
|
Temp |
98°C |
98°C |
64°C |
72°C |
98°C |
57°C |
72°C |
72°C |
4°C |
|
Time |
5:00 |
0:15 |
0:30 |
2:00 |
0:15 |
0:30 |
2:00 |
5:00 |
Hold |
15 cycles 15 cycles
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.4g of agarose and 50 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 50 ul PCR product + 10 ul 6X gel loading buffer.
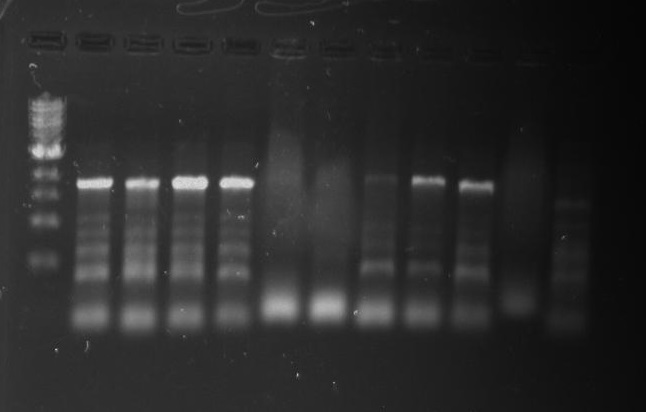
Gel 27: From left to right. Ladder, 326 DivJ, Minimal DivJ, pXYFPC-2, pYCFPC-4.
-start overnight cultures CA
Lab Notebook 9/12/13
Miniprep of Overnight CA8, CA9, YD1 and YD3 Cultures:
Spin down overnight culture for 10 minutes at 3,000 xg. Discard 3.4 ml of the supernatant. Resuspend the pellet.
1) Add 100 ul of 7X Lysis buffer to 600 ul of e. coli culture in a 1.5 ml microcentrifuge tube. Mix by inverting the tube 4-6 times and lyse samples for 1-2 minutes.
2) Add 350 ul of cold Neutralization buffer. Mix thoroughly. Neutralization is complete when sample becomes yellow and precipitate has formed.
3) Centrifuge at 16,000 xg for 2 minutes.
4) Transfer the supernatant into the Zymo-Spin IIN column .
5) Place column into a collection tube and centrifuge at 11,000 xg for 15 seconds. Discard the flow through and place the column back into the same collection tube.
6) Add 200 ul of Endo Wash buffer to the column. Centrifuge at 11,000 xg for 30 seconds.
7) Add 400 ul of Zyppy Wash buffer to the column. Centrifuge at 11,000 xg for 30 seconds.
8) Transfer the column into a clean 1.5 ml microcentrifuge tube then add 30 ul of MilliQ water. Let stand for one minute at room temperature. Centrifuge at 11,000 xg for 15 seconds to elute the DNA.
Purification of DivJ:
1) Cut band out in a dark room with a sterile razor and an UV lamp.
2) Place the cut of gel band in a
3) Centrifuge the gel band for 10 minutes at 5,000 g.
4) Add five volumes of DNA Binding buffer to each volume of DNA sample.
5) Load the mixture into a Zymo-Spin Column in a collection tube.
6) Centrifuge at 10,000 g for 30 seconds. Discard flow through.
7) Add 200 ul of DNA Wash buffer to the column and centrifuge at 10,000 g for 30 seconds.
8) Place the Zymo-Spin column into a new 1.5 ml tube.
9) Add 12 ul MilliQ water to column matrix and spin at 10,000 g for 30 seconds to elute the DNA.
Restriction Digest of DivJ:
|
Minimal DivJ |
326 DivJ |
pXYFPC-2 |
pYCFPC-4 |
|
28 ul DNA |
28 ul DNA |
3.5 ul DNA |
3.5 ul DNA |
|
5 ul 10X CutSmart buffer |
5 ul 10X CutSmart buffer |
5 ul 10X CutSmart buffer |
5 ul 10X CutSmart buffer |
|
1 ul AciI |
1 ul AciI |
1 ul AciI |
1 ul AciI |
|
1 ul NdeI |
1 ul NdeI |
1 ul NdeI |
1 ul NdeI |
|
15 ul MilliQ water |
15 ul MilliQ water |
39.5 ul MilliQ water |
39.5 ul MilliQ water |
Incubate at 37°C for 30 minutes.
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.4g of agarose and 50 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 50 ul PCR product + 10 ul 6X gel loading buffer.
Gel 28: From left ro right. Ladder, pXYFPC-2, pYCFPC-4.
Lab Notebook 9/13/13
Clean and concentrated DivJ and eluted it to 12 ul. Cut out the vectors. Clean and concentrated and eluted it to 15 ul each.
Ligation of DivJ:
|
Mastermix of pXYFPC-2 |
Mastermix of pYCFPC-4 |
|
12 ul pXYFPC-2 |
12 ul pYCFPC-4 |
|
3 ul 10X T4 buffer |
3 ul 10X T4 buffer |
|
1.5 ul T4 Ligase |
1.5 ul T4 Ligase |
Six total ligation reactions:
|
1) 5.5 ul of Mastermix for pXYFPC-2 + 4.5 ul of 326 DivJ |
4) 5.5 ul of Mastermix for pVCPFPC-4 + 4.5 ul of 326 DivJ |
|
2) 5.5 ul of Mastermix for pXYFPC-2 + 4.5 ul of MilliQ water |
5) 5.5 ul of Mastermix for pVCPFPC-4 + 4.5 ul of MilliQ water |
|
3) 5.5 ul of Mastermix for pXYFPC-2 + 4.5 ul of Minimal DivJ |
6) 5.5 ul of Mastermix for pVCPFPC-4 + 4.5 ul of Minimal DivJ |
Incubate ligation reactions for 30 minutes or greater at room temperature.
Heat Shock Transformations:
1) Add 100 ul of Top10 Chemicompetent cells in 1-2 ul of each ligation reaction.
2) Place in a 42°C water bath for 30 seconds.
3) Place back on ice for 5 minutes.
4) Add 500 ul of SOC to a microcentrifuge tube.
5) Transfer the Top10 Chemicompetent cells and ligation reaction to the microcentrifuge tube.
6) Incubate for one hour at 37°C at 200 rpm.
Plating the Transformations:
1) Obtain three plates of LB and gentamicin and three plates of LB and kanamycin.
2) Warm up the plates in the 37°C prior to plating.
3) Pipet all of the transformation into the plate.
4) Using a bunsen burner, heat up the steel rod.
5) Create streaks with the steel rod.
6) Wait for 5 minutes until all the liquid is soaked up by the plate.
7) Tape up the plates and/or place the plates in a plastic bag.
8) Place plates in an incubator at 37°C.
Lab Notebook 9/14/13
Colony PCR of TipF and DivJ Plates:
Mastermix for pVCPFPC-4 plasmids:
- 110 ul 2x OneTaq
- 44 ul MilliQ water
- 22 ul Pvan-for primer at 10 uM concentration
- 22 ul eGYC-1 primer at 10 uM concentration
Mastermix for pXYFPC-2 plasmids:
- 55 ul 2x OneTaq
- 22 ul MilliQ water
- 11 ul Pxyl-for primer at 10 uM concentration
- 11 ul eGYC-1 primer at 10 uM concentration
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 10 ul PCR product + 2 ul 6X gel loading buffer.
Gel 29: From left to right. Top: CD1, CD2, CD3, CD4,CD5, CD6, CD7, CD8, CD9, CD10 CA1, CA2, CA3, CA4, CA5, CA6, CA7, CA8, Positive Control for pVCPFPC-4. Bottom: CA10, Positive Control for pVCPFPC-4, YA1, YA2, YA3, YA4, YA5, YA6, YA7, YA8. YA9, YA10, Positive Control for pXYFPC-2. CD = 326 DivJ in pVCPFPC-4. CA = Minimal DivJ in pVCPFPC-4. YA = Minimal DivJ in pXYFPC-2.
Lab Notebook 9/15/13
Overnight Cultures of YA1, YA2, YA3, YA4, YA5, CA5, CA10, CD2, CD3:
For each overnight:
3 ml of SOC. For pVCPFPC-4, add in 4.5 ul of gentamicin. For pXYFPC-2, add in 1.8 ul of kanamycin.
Lab Notebook 9/16/13
Miniprep of Overnight Cultures:
1) Add 100 ul of 7X Lysis buffer to 600 ul of e. coli culture in a 1.5 ml microcentrifuge tube. Mix by inverting the tube 4-6 times and lyse samples for 1-2 minutes.
2) Add 350 ul of cold Neutralization buffer. Mix thoroughly. Neutralization is complete when sample becomes yellow and precipitate has formed.
3) Centrifuge at 16,000 xg for 2 minutes.
4) Transfer the supernatant into the Zymo-Spin IIN column .
5) Place column into a collection tube and centrifuge at 11,000 xg for 15 seconds. Discard the flow through and place the column back into the same collection tube.
6) Add 200 ul of ENdo Wash buffer to the column. Centrifuge at 11,000 xg for 30 seconds.
7) Add 400 ul of Zyppy Wash buffer to the column. Centrifuge at 11,000 xg for 30 seconds.
8) Transfer the column into a clean 1.5 ml microcentrifuge tube then add 30 ul of MilliQ water. Let stand for one minute at room temperature. Centrifuge at 11,000 xg for 15 seconds to elute the DNA.
Nine Sequencing Samples:
YA1, YA2, YA3, YA4, YA5, CA5, CA10, CD2, CD3
10 ul of miniprepped DNA plasmid with 3 ul of the resepective forward/reverse primer at 1 uM concentration.
Lab Notebook 9/17/13
Made more gentamicin and kanamycin plates.
Re-did the synthesis of the minimal DivJ 60 mers (sequencing results suggest the first DivJM assembly resulted in a single base deletion.
Transformation of the CA8 and CA9 miniprep plasmid that we sent in for sequencing. Changed the amount of SOC added. This time we used 250 ul.
Lab Notebook 9/18/13
Re-did the restriction digest of the vectors and the minimal DivJ with the restriction enzymes NdeI and AciI.
Lab Notebook 9/19/13
Re-did the colony PCR of the TipF plates one last time. Same settings as before.
Lab Notebook 9/20/13
Colony PCR of the Minimal DivJ Plate:
Mastermix for pVCPFPC-4 plasmids:
- 60 ul 2x OneTaq
- 24 ul MilliQ water
- 12 ul Pvan-for primer at 10 uM concentration
- 12 ul eGYC-1 primer at 10 uM concentration
Mastermix for pXYFPC-2 plasmids:
- 60 ul 2x OneTaq
- 24 ul MilliQ water
- 12 ul Pxyl-for primer at 10 uM concentration
- 12 ul eGYC-1 primer at 10 uM concentration
Touchdown PCR Protocol:
|
Temp |
94°C |
94°C |
60°C |
68°C |
94°C |
53°C |
68°C |
68°C |
4°C |
|
Time |
5:00 |
0:15 |
0:30 |
0:30 |
0:15 |
0:30 |
0:30 |
5:00 |
Hold |
15 cycles 15 cycles
 "
"