02/08/13
From 2013.igem.org
(Difference between revisions)
| (5 intermediate revisions not shown) | |||
| Line 37: | Line 37: | ||
*the plates with the ligated DNA did not show growth as the RFP plates, this could be because the cells did not take up as much plasmid as the optimum RFP plasmid or not all the plasmid were ligated with the biobrick. | *the plates with the ligated DNA did not show growth as the RFP plates, this could be because the cells did not take up as much plasmid as the optimum RFP plasmid or not all the plasmid were ligated with the biobrick. | ||
*It is also important to notice that there is not much different in the number of colonies between the 5ul and 10ul plates. | *It is also important to notice that there is not much different in the number of colonies between the 5ul and 10ul plates. | ||
| + | |||
| + | ==Next steps== | ||
| + | * The presence of the limonene biobrick in the ligated product has to be confirmed. | ||
| + | * This would be done in 2 ways | ||
| + | a) Measurement of volatile organic compounds (VOC) - 3 separate groups of cells would be prepared for this experiment. | ||
| + | * The first group would be transformed cells containing the ligated product. | ||
| + | * The second group would be transformed cells containing only circularised pSB1C3 vector | ||
| + | * The third group would just be competent cells | ||
| + | b) Miniprep of transformed cells containing the ligated product (from above results) | ||
| + | |||
==Digestion of pSB1C3 backbone== | ==Digestion of pSB1C3 backbone== | ||
*Master Mix (changes were made from the protocol) | *Master Mix (changes were made from the protocol) | ||
| Line 54: | Line 64: | ||
*Add the following substances | *Add the following substances | ||
**12ul of dH2O | **12ul of dH2O | ||
| - | **1ul of QS Ligase * | + | **1ul of QS Ligase* |
**2ul of the digested vector pSB1C3 | **2ul of the digested vector pSB1C3 | ||
**5ul of 4xQS Buffer (vortex before use) | **5ul of 4xQS Buffer (vortex before use) | ||
| Line 63: | Line 73: | ||
*For the control, no QS Ligase is added but its equivalent volume (1ul) is made up by dH2O. | *For the control, no QS Ligase is added but its equivalent volume (1ul) is made up by dH2O. | ||
| + | |||
| + | ==Gel Electrophoresis== | ||
| + | * Gel was run in order to confirm that the cirularisation experiment had worked. | ||
| + | * The linearised pSB1C3 vector was used as a control. | ||
| + | * The results showed that the circularisation experiment worked, as the linearised vector is represented on the gel while the circularised vector was cloudy and ran off the gel. | ||
| + | |||
| + | ==Transformation of circularised pSB1C3 vector == | ||
| + | * After confirming the presence of the circularised vector, it was transformed into competent cells | ||
| + | * This was left to incubate overnight at 37 degrees Celsius | ||
| + | |||
| + | |||
| + | ==Reculturing of transformed cells == | ||
| + | * The transformed cells containing the ligated product from yesterday's experiment were recultured onto 2 agar plates. | ||
| + | * The 2 plates were divided into 12 squares each. | ||
| + | * Each square was streaked with colonies that were randomly chosen from the 5ul and 10ul plates. | ||
| + | * The plates were then left on the bench for overnight incubation | ||
| + | |||
| + | ==Incubation in broth== | ||
| + | * The next day the some colonies from the recultured plates were incubated in broth. | ||
| + | * This was done in preparation of the miniprep. | ||
Latest revision as of 13:20, 6 August 2013
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Modeling | Notebook | Safety | Attributions |
|---|
Contents |
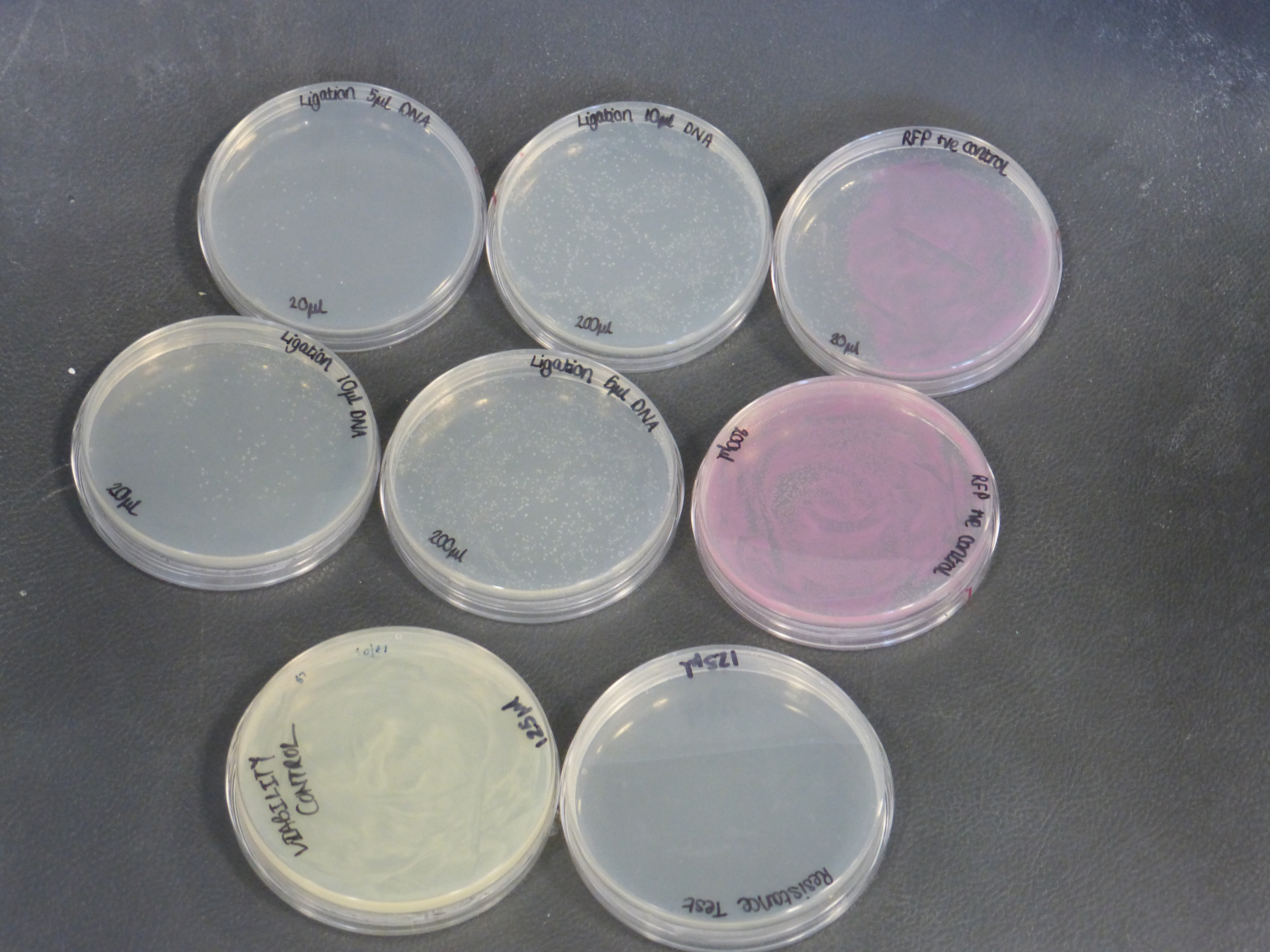
Results from the transformation plates
- On the 01/08/2013 we plated the transformations containing the biobrick BBa_K118025.
- Our transformations were successful
- the plates contained different volumes of the transformation: 20ul and 200ul
- the plate to test viability shows growth as expected
- the plate to test resistance shows no growth as expected
- there were two different different transformations that contained different volumes of ligated DNA, which were plated into different plates.
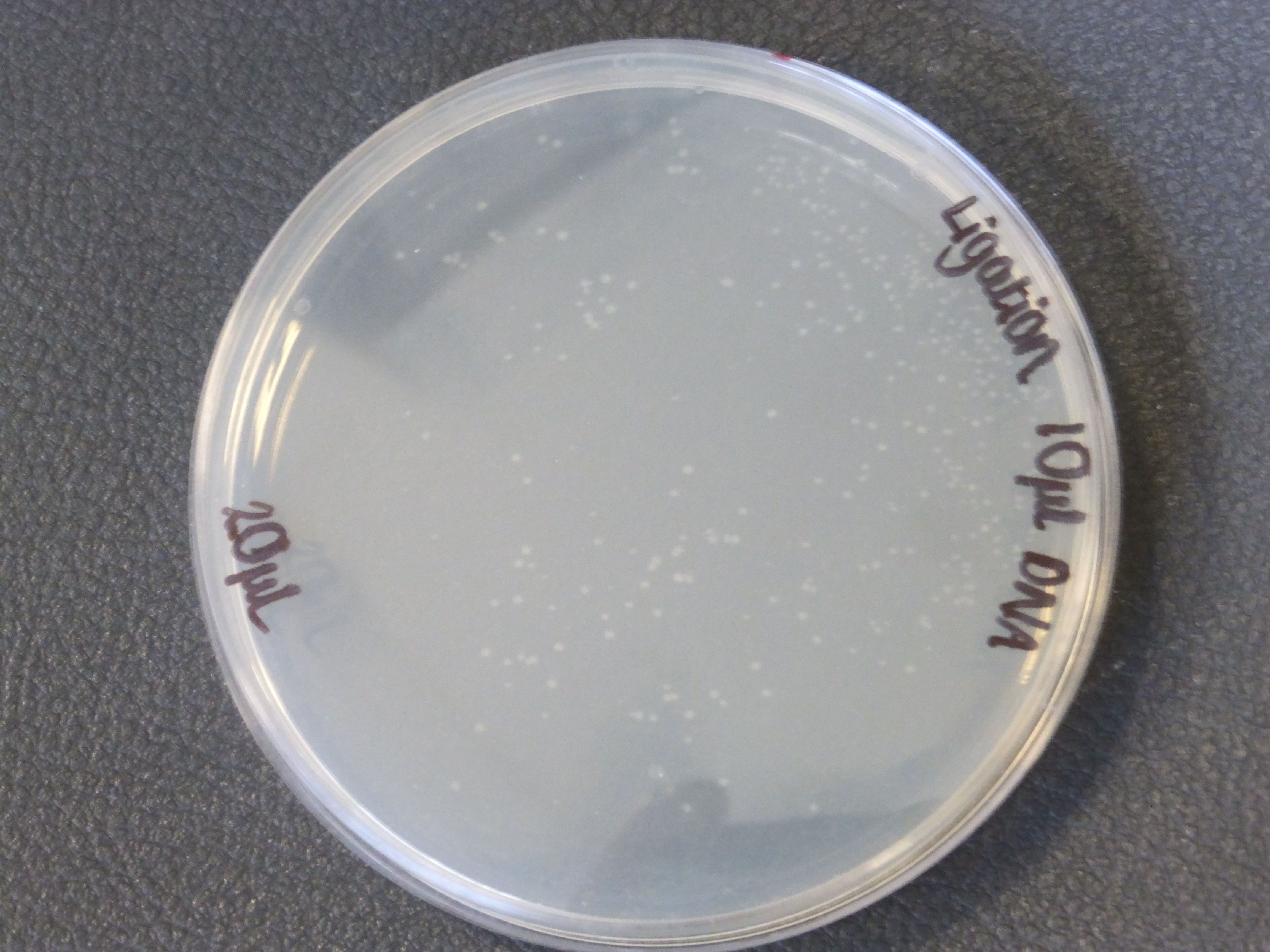
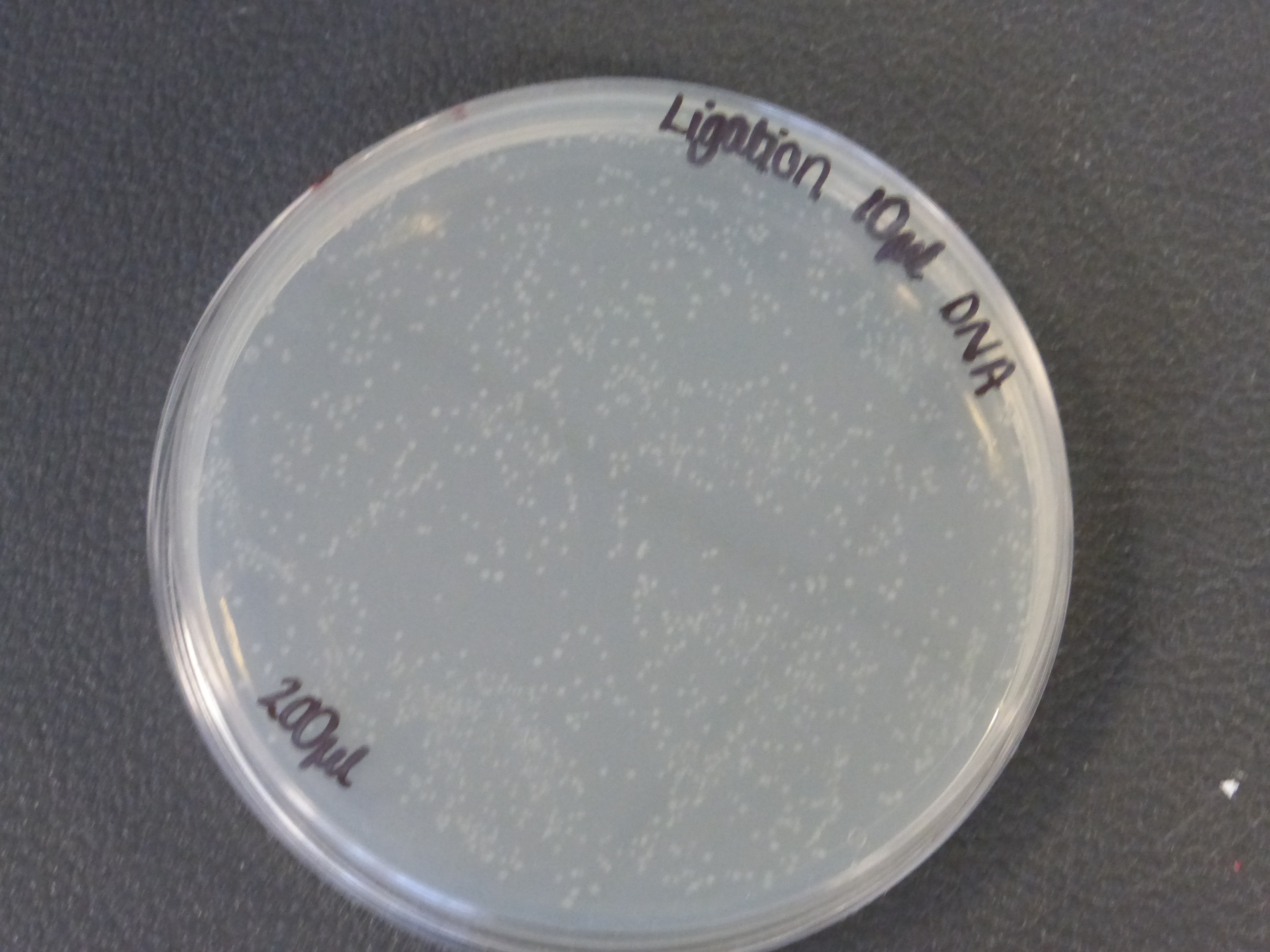
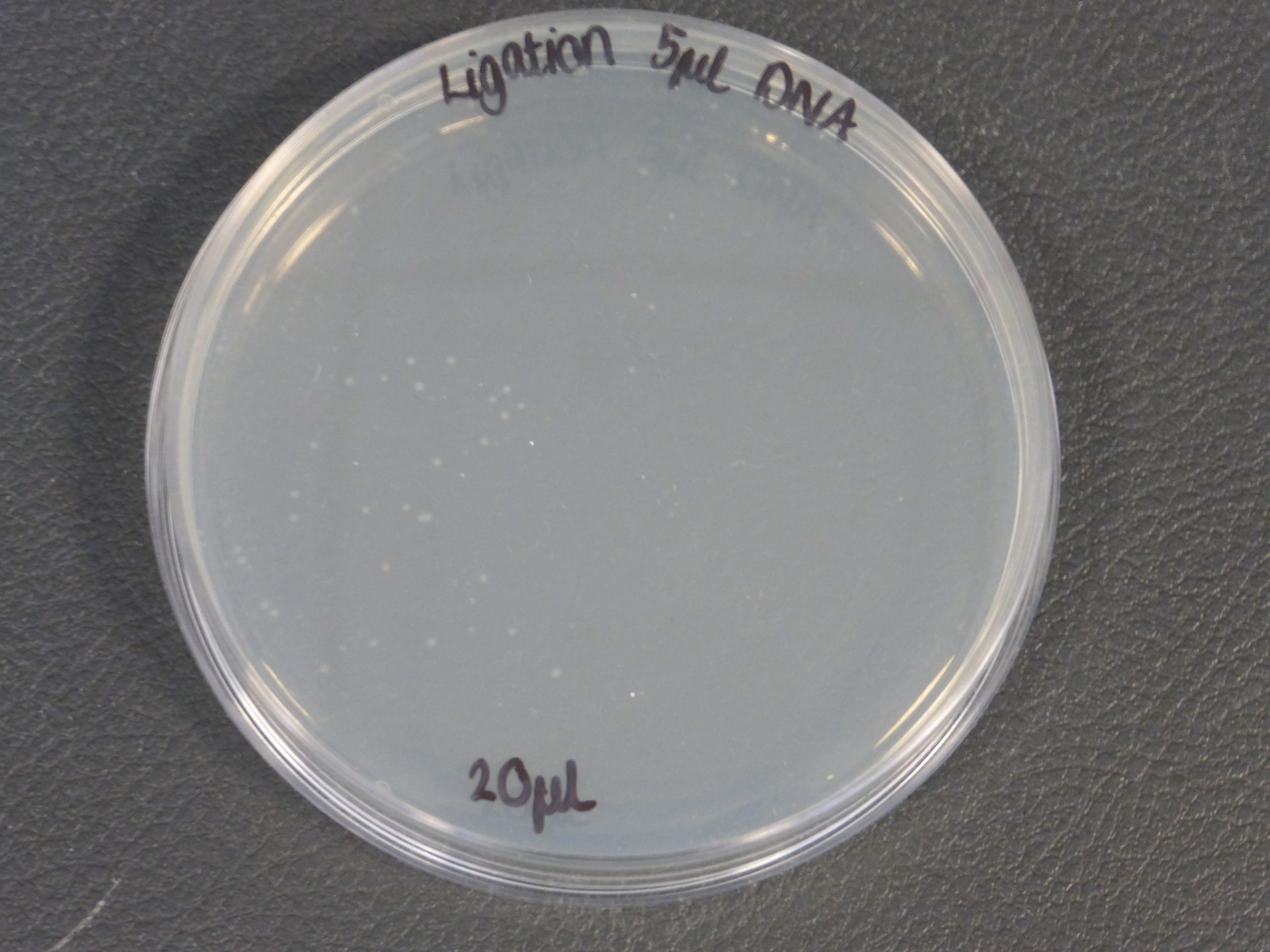
- The 10ul ligated DNA plates showed growth
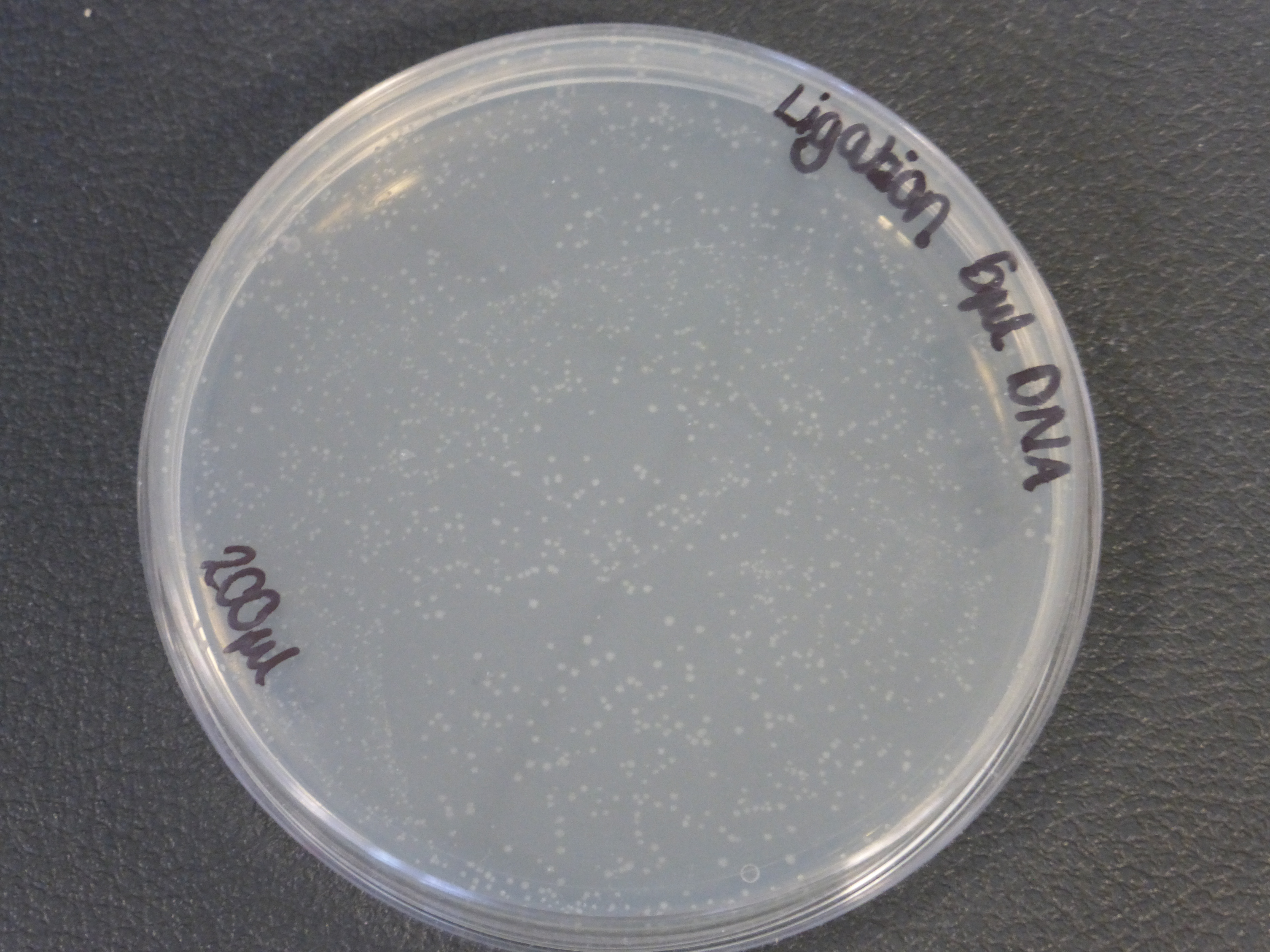
- The 5ul ligated DNA plates showed growth
- The RFP control plates showed growth as expected
- the plates with the ligated DNA did not show growth as the RFP plates, this could be because the cells did not take up as much plasmid as the optimum RFP plasmid or not all the plasmid were ligated with the biobrick.
- It is also important to notice that there is not much different in the number of colonies between the 5ul and 10ul plates.
Next steps
- The presence of the limonene biobrick in the ligated product has to be confirmed.
- This would be done in 2 ways
a) Measurement of volatile organic compounds (VOC) - 3 separate groups of cells would be prepared for this experiment.
- The first group would be transformed cells containing the ligated product.
- The second group would be transformed cells containing only circularised pSB1C3 vector
- The third group would just be competent cells
b) Miniprep of transformed cells containing the ligated product (from above results)
Digestion of pSB1C3 backbone
- Master Mix (changes were made from the protocol)
- 5ul cut smart Buffer
- 0.5ul of XbaI
- 0.5ul of SpeI
- 19ul of dH2O
- For each reaction add:
- 4ul of linearized backbone
- 4ul of enzyme master mix
- digest at 37C for 30min
- heat kill at 80C for 20min
- this digestion was done as control to test if our colonies with the limonene biobrick is expressing limonene.
Circularisation of pSB1C3
- For this experiment, two separate samples will be run; a control and the actual ligation.
- Add the following substances
- 12ul of dH2O
- 1ul of QS Ligase*
- 2ul of the digested vector pSB1C3
- 5ul of 4xQS Buffer (vortex before use)
- Mix thoroughly by pipetting
- Incubate at room temperature for 5 min to create cohesive ends
- Run 2.5-5ul of the ligation mixture onto an agarose gel to check ligation efficiency against a known marker. Add 5ul of dye to aid the visualization of results.
- Transform it into competent cells (E.coli)
*For the control, no QS Ligase is added but its equivalent volume (1ul) is made up by dH2O.
Gel Electrophoresis
- Gel was run in order to confirm that the cirularisation experiment had worked.
- The linearised pSB1C3 vector was used as a control.
- The results showed that the circularisation experiment worked, as the linearised vector is represented on the gel while the circularised vector was cloudy and ran off the gel.
Transformation of circularised pSB1C3 vector
- After confirming the presence of the circularised vector, it was transformed into competent cells
- This was left to incubate overnight at 37 degrees Celsius
Reculturing of transformed cells
- The transformed cells containing the ligated product from yesterday's experiment were recultured onto 2 agar plates.
- The 2 plates were divided into 12 squares each.
- Each square was streaked with colonies that were randomly chosen from the 5ul and 10ul plates.
- The plates were then left on the bench for overnight incubation
Incubation in broth
- The next day the some colonies from the recultured plates were incubated in broth.
- This was done in preparation of the miniprep.
 "
"