05/08/13
From 2013.igem.org
(Difference between revisions)
| (7 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | <div style="font-family:arial;padding:5px;border-radius:5px;border:5px solid #FF2800;"> | ||
| + | {| style="color:#87EA00;background-color:#FFFFFF;" cellpadding="2" cellspacing="2" border="0" bordercolor="#000000" width="100%" align="center" | ||
| + | !align="center"|[[Team:Leicester|Home]] | ||
| + | !align="center"|[[Team:Leicester/Team|Team]] | ||
| + | !align="center"|[https://igem.org/Team.cgi?year=2013&team_name=Leicester Official Team Profile] | ||
| + | !align="center"|[[Team:Leicester/Project|Project]] | ||
| + | !align="center"|[[Team:Leicester/Parts|Parts Submitted to the Registry]] | ||
| + | !align="center"|[[Team:Leicester/Modeling|Modeling]] | ||
| + | !align="center"|[[Team:Leicester/Notebook|Notebook]] | ||
| + | !align="center"|[[Team:Leicester/Safety|Safety]] | ||
| + | !align="center"|[[Team:Leicester/Attributions|Attributions]] | ||
| + | |} | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <p> | ||
| + | |||
==Isolation of ligated plasmid== | ==Isolation of ligated plasmid== | ||
*The ligated plasmid- chloramphenicol backbone and limonene biobrick, were isolated from cells that were incubated overnight in broth by miniprep. | *The ligated plasmid- chloramphenicol backbone and limonene biobrick, were isolated from cells that were incubated overnight in broth by miniprep. | ||
* Protocols from both Bioline kit and Thermoscientific were used for the miniprep. | * Protocols from both Bioline kit and Thermoscientific were used for the miniprep. | ||
* there were 6 tubes containing the plasmid. | * there were 6 tubes containing the plasmid. | ||
| - | * The plasmid concentration in each of the 6 tubes were measured. | + | * The plasmid concentration in each of the 6 tubes were measured using nanodrop. |
* 200ng of plasmid from each of the 6 different tube was used for digestion. | * 200ng of plasmid from each of the 6 different tube was used for digestion. | ||
| + | |||
| + | {|border=1 | ||
| + | |sample||volume(ul)||concentration (ng/ul)||yield(ug) | ||
| + | |- | ||
| + | |5.1||99||75||7.4 | ||
| + | |- | ||
| + | |5.2||100||44.2||4.4 | ||
| + | |- | ||
| + | |5.3||98||46.7||4.6 | ||
| + | |- | ||
| + | |10.1||46||61.8||2.84 | ||
| + | |- | ||
| + | |10.2||44||77.1||3.39 | ||
| + | |- | ||
| + | |10.3||47||13.7||0.64 | ||
| + | |} | ||
==Digestion of ligated plasmid== | ==Digestion of ligated plasmid== | ||
| Line 17: | Line 50: | ||
*The product of digestion was run on agarose gel | *The product of digestion was run on agarose gel | ||
**This was done in order to confirm the presence of the limonene biobrick | **This was done in order to confirm the presence of the limonene biobrick | ||
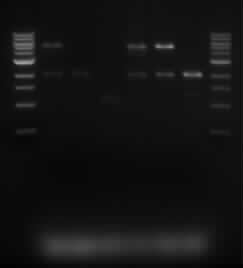
| + | [[File:iGem_Digested_050813.jpg]] | ||
| + | *The lanes are in the following order: | ||
| + | **Marker, Sample 5.1, Sample 5.2, Sample 5.3, Sample 10.1, Sample 10.2, Sample 10.3, Marker. | ||
| + | *We were expecting to see a 2kb and 5kb band. These bands appeared for samples 5.1, 10.1,10.2 and 10.3. To further determine the composition of samples, we did a single digest on the samples. | ||
| + | |||
| + | ==Single digest== | ||
| + | *For samples 5.1, 10.1, 10.2 and 10.3 two single digests each were made with EcoRI and PstI, giving a total of 8 samples. | ||
| + | *The samples are run on 0.8% agarose gel. | ||
Latest revision as of 12:30, 6 August 2013
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Modeling | Notebook | Safety | Attributions |
|---|
Contents |
Isolation of ligated plasmid
- The ligated plasmid- chloramphenicol backbone and limonene biobrick, were isolated from cells that were incubated overnight in broth by miniprep.
- Protocols from both Bioline kit and Thermoscientific were used for the miniprep.
- there were 6 tubes containing the plasmid.
- The plasmid concentration in each of the 6 tubes were measured using nanodrop.
- 200ng of plasmid from each of the 6 different tube was used for digestion.
| sample | volume(ul) | concentration (ng/ul) | yield(ug) |
| 5.1 | 99 | 75 | 7.4 |
| 5.2 | 100 | 44.2 | 4.4 |
| 5.3 | 98 | 46.7 | 4.6 |
| 10.1 | 46 | 61.8 | 2.84 |
| 10.2 | 44 | 77.1 | 3.39 |
| 10.3 | 47 | 13.7 | 0.64 |
Digestion of ligated plasmid
- Master Mix (changes were made from the protocol)
- 2ul of NEB Buffer
- 1ul of EcoRI
- 1ul of PstI
- varying concentration of dH2O and plasmid depending on overall concentration of plasmid in each tube.
- Final volume of each of the 6 tubes was 20ul.
Gel Electrophoresis
- The product of digestion was run on agarose gel
- This was done in order to confirm the presence of the limonene biobrick
- The lanes are in the following order:
- Marker, Sample 5.1, Sample 5.2, Sample 5.3, Sample 10.1, Sample 10.2, Sample 10.3, Marker.
- We were expecting to see a 2kb and 5kb band. These bands appeared for samples 5.1, 10.1,10.2 and 10.3. To further determine the composition of samples, we did a single digest on the samples.
Single digest
- For samples 5.1, 10.1, 10.2 and 10.3 two single digests each were made with EcoRI and PstI, giving a total of 8 samples.
- The samples are run on 0.8% agarose gel.
 "
"