Team:GeorgiaTech/Human Practices
From 2013.igem.org
Henry54809 (Talk | contribs) |
Henry54809 (Talk | contribs) |
||
| Line 1: | Line 1: | ||
{{Team:GeorgiaTech/template}} | {{Team:GeorgiaTech/template}} | ||
| + | |||
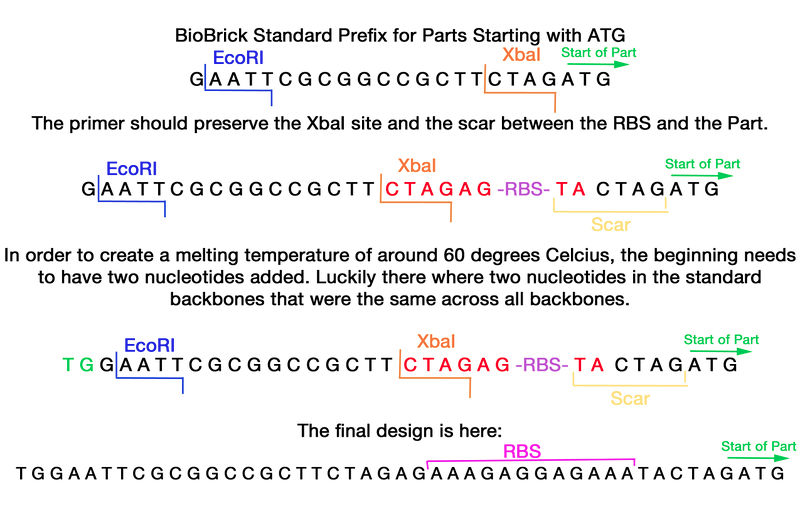
For the duration of the summer, the biggest question our team struggled with was: how can we make our experiments and cloning projects more efficient? Organisms take time to grow; reactions can take hours to complete. When experiments fail, human resources, money and time are utilized. We found a particular challenge in the incorporation of small (~tens of base pairs) fragments, such as the ribosomal binding site (RBS), into existing BioBrick parts. Most BioBrick parts require the addition of a RBS to express the protein. We found the use of standard or 3A assembly extremely inefficient in assembling RBS with BioBrick parts, and this created a significant bottleneck in our projects’ progress. Our goal was to invent a faster, more efficient method of assembling small sequences, including RBS as our trial candidate, into existing parts compatible with the BioBrick registry. | For the duration of the summer, the biggest question our team struggled with was: how can we make our experiments and cloning projects more efficient? Organisms take time to grow; reactions can take hours to complete. When experiments fail, human resources, money and time are utilized. We found a particular challenge in the incorporation of small (~tens of base pairs) fragments, such as the ribosomal binding site (RBS), into existing BioBrick parts. Most BioBrick parts require the addition of a RBS to express the protein. We found the use of standard or 3A assembly extremely inefficient in assembling RBS with BioBrick parts, and this created a significant bottleneck in our projects’ progress. Our goal was to invent a faster, more efficient method of assembling small sequences, including RBS as our trial candidate, into existing parts compatible with the BioBrick registry. | ||
Revision as of 00:13, 28 September 2013
Template:Team:GeorgiaTech/template
For the duration of the summer, the biggest question our team struggled with was: how can we make our experiments and cloning projects more efficient? Organisms take time to grow; reactions can take hours to complete. When experiments fail, human resources, money and time are utilized. We found a particular challenge in the incorporation of small (~tens of base pairs) fragments, such as the ribosomal binding site (RBS), into existing BioBrick parts. Most BioBrick parts require the addition of a RBS to express the protein. We found the use of standard or 3A assembly extremely inefficient in assembling RBS with BioBrick parts, and this created a significant bottleneck in our projects’ progress. Our goal was to invent a faster, more efficient method of assembling small sequences, including RBS as our trial candidate, into existing parts compatible with the BioBrick registry.
After brainstorming a bit, we had an idea: what if during a simple PCR amplification, we could add the RBS BioBrick BBa_B0034 with the right primers? By doing so, the time taken to add RBS to any BioBrick would be reduced to half, and consist of 2 hour PCR reactions and a ligating stage onto a backbone. An easy oligo primer, that could be used in PCR extension and then placed into a vector backbone, would save time, resources, and frustration. The RBS primer is a standard primer that can be used for site directed mutagenesis in all linearized backbones available from the iGEM database
By running the PCR reaction with these primers, our team saw significant amplification of nearly every BioBrick part that fit our qualifications. This is remarkable since the ATG site was a small site to attach to the part after the ribosomal binding site. After the creation of the BioBrick with the RBS, our project and its progress made significant leaps, moving much faster than we had before. It was a great addition not only to our individual project, but to the synthetic biology community as a whole.
This approach could launch a new era of synthetic biology that would not need restriction enzymes and ligase for every combination of small parts, but rather a simple primer and a PCR amplification reaction. The next step would be to determine whether a promoter region could be added on in a secondary PCR reaction, obtaining a fully functional part, ready for use and testing in any competent E. coli site. This new practice in synthetic biology could reduce the amount of time needed to conduct experiments, and we could make advances in research in a greater amount of time than we ever have before.
 "
"