Team:Heidelberg/Templates/Indigoidine week10
From 2013.igem.org
(Created page with " In parallel to the experiments with the pMM-plasmids (Konrad) we start to get the native bpsA from Streptomyces lavendulae lavendulae DSMXXXX (Ralf). ===Indigoidine productio...") |
|||
| Line 10: | Line 10: | ||
==== analytic digestion ==== | ==== analytic digestion ==== | ||
==== fragment amplification ==== | ==== fragment amplification ==== | ||
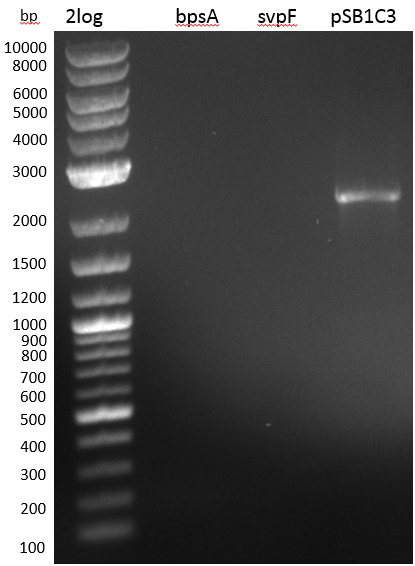
| - | [[File: | + | [[File:Heidelberg_2013-07-02 DelHf1a DelHf2 M bpsA pSB1C3.jpg|500px|thumb|right|PCR for bpsA and backbone amplification and |
Phusion polymerase validation (lanes to the right side of ladder; white stripe on the right is not a band); | Phusion polymerase validation (lanes to the right side of ladder; white stripe on the right is not a band); | ||
| Line 122: | Line 122: | ||
====Agarose Gel==== | ====Agarose Gel==== | ||
| - | [[ | + | [[File:Heidelberg_20130705_Slav.jpg|300px|gel with 17 ul PCR product, 3 ul 6x buffer, 6 ul 2-log]] |
* pSB1C3 backbone | * pSB1C3 backbone | ||
** weak band on gel < 50 ng/ 17 ul | ** weak band on gel < 50 ng/ 17 ul | ||
| Line 204: | Line 204: | ||
0,8 % Agarose Gel | 0,8 % Agarose Gel | ||
| - | :[[ | + | :[[File:Heidelberg_20130705_slav2.PNG|300px|2nd run PCR for RB construct 1]] |
* pSB1C3 worked well with 20-50 ng/ ul | * pSB1C3 worked well with 20-50 ng/ ul | ||
* BpsA | * BpsA | ||
| Line 288: | Line 288: | ||
Desperation Gel | Desperation Gel | ||
| - | :[[ | + | :[[File:Heidelberg_20130705_slav3.jpg|500px|0,8 % Agarose, 100 V, 60 min.]] |
* band around 200 bp? unspecific product due to PCR conditions, but then why no product? | * band around 200 bp? unspecific product due to PCR conditions, but then why no product? | ||
* VI glowing pocket -> sucrose in YEME medium? -> PCR purification and new gel -> still nothing | * VI glowing pocket -> sucrose in YEME medium? -> PCR purification and new gel -> still nothing | ||
| Line 331: | Line 331: | ||
Gel 0,8 % 100 V 60 min | Gel 0,8 % 100 V 60 min | ||
| - | :[[ | + | :[[File:Heidelberg_20130706_svp5.PNG|400 px|PCR with RB-Primers and intron iTaq]] |
*nothing | *nothing | ||
** intron iTaq maybe doesn't work so well since control isn't working; or standard protocol is bad. | ** intron iTaq maybe doesn't work so well since control isn't working; or standard protocol is bad. | ||
| Line 373: | Line 373: | ||
<gallery widths="500px" heights="350px"> | <gallery widths="500px" heights="350px"> | ||
| - | File: | + | File:Heidelberg_20130706_svp6_1.PNG|500px|2-log; A-E; 2-log; F-J |
| - | File: | + | File:Heidelberg_20130706_svp6_2.PNG|2-log; K-O; 2-log; Slav IVb; VI; V; VII |
</gallery> | </gallery> | ||
| Line 405: | Line 405: | ||
|} | |} | ||
gel 9 ul 1x loading buffer, 1 ul pcr product | gel 9 ul 1x loading buffer, 1 ul pcr product | ||
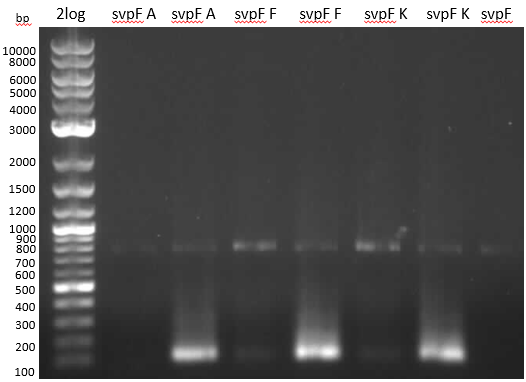
| - | :[[ | + | :[[File:Heidelberg_20130707_svp7.PNG|400px|2-log / A / A-Phu Fl / F / F-Phu-Fl / K / K-Phu-Fl / svp 7,2 ng template]] |
*gel analysis | *gel analysis | ||
** no significant amplification of svp with Phusion Flash | ** no significant amplification of svp with Phusion Flash | ||
| Line 443: | Line 443: | ||
|} | |} | ||
gel with 9 ul 1x loading buffer and 1 ul pcr product | gel with 9 ul 1x loading buffer and 1 ul pcr product | ||
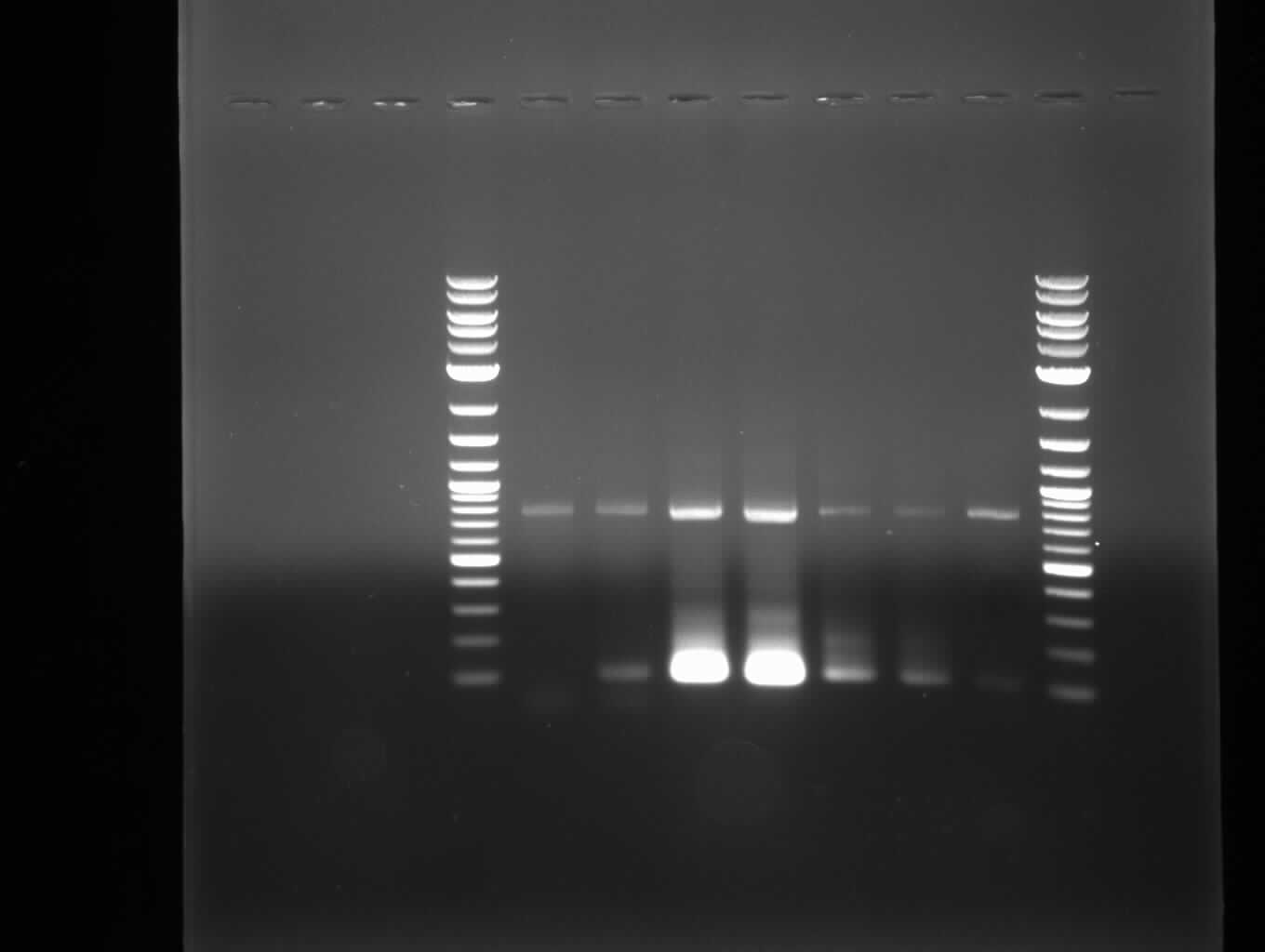
| - | :[[ | + | :[[File:Heidelberg_20130707_svp8.JPG|400px|2-log / wrong / wrong / 3-step K / 3-step K-Phu-Fl / 2-step K / 2-step K-Phu-Fl / K |
template / 2-log]] | template / 2-log]] | ||
Latest revision as of 00:44, 5 October 2013
In parallel to the experiments with the pMM-plasmids (Konrad) we start to get the native bpsA from Streptomyces
lavendulae lavendulae DSMXXXX (Ralf).
Contents |
Indigoidine production with pKH1 (Konrad)
colony PCR
transformation
analytic digestion
fragment amplification
==> for two fragments with our Phusion MM and Phusion MM of SYNtheSYS (old, don't know if functional...)
- 0.4 µl template
- 2x 0.5 µl Primer
- 25 µl Phusion MM
- 14.6 µl H2O
| fragment | primer | template (DNA) | annealing temp (X1;X2) [°C] | elongation time (Y) [s] |
|---|---|---|---|---|
| f1: bpsA (all) | (NI01,NI06) | pMM64 | 65;66 | 120 |
| f7: pSB1C3 (linear.) | (NI09,NI10) | pSB1C3 with J04450 (pJM03) | 66;61 | 75 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 10 | 98 | 5 |
| X1 (incr. down with 0.5 °C) | 15 | |
| 72 | Y | |
| 20 | 98 | 5 |
| X2 | 15 | |
| 72 | Y | |
| 1 | 72 | 360 |
| 1 | 4 | inf |
- RESULT: no PCR product for both amplicons, maybe due to wrong cycle conditions
construction of pSB1C3-bpsA-svp pRB1
Streptomyces lavendulae lavendulae DSMZXXXX was cultivated in GYM medium from freeze dried cell pellet at 28 °C and 170 rpm. cultivation of S. lavendulae lavendulae
- freeze dried cell pellet in 5 ml medium 65
- 3x 200 ul in 3 mL medium 65 and YEME, respectively. 28 °C 200 rpm (3 p.m.)
- 3x agar plate medium 65, 50/75/100 ul inoculation; 28 °C (7 p.m.)
- prepare YEME agar for next day
PCR with Gibson Primers RB05-RB10
- pSB1C3 backbone from PCR product 50 uL Phusion Flash; 05 uM Primer, 25 uL Master mix, 19,4 uL water, 0,6 uL pSB1C3
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72 | 40 | |
| 1 | 72 | 420 |
| 1 | 4 | inf Gibson assembly and |
- BpsA colony PCR 50 ul Phusion Flash; 0,5 uM Primer, 25 uL Master Mix, 20 uL colony with water
- colony was picked and held into liquid nitrogen/ warm water repeatedly
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 11 |
| 1 | 65 | 5 |
| 1 | 72 | 60 |
| 30 | 98 | 1 |
| 72 | 60 | |
| 1 | 72 | 300 |
| 1 | 4 | inf |
- pMM65 PCR for svp 50 ul Phusion Flash; 0,5 uM Primer, 25 uL Master mix, 15 uL water, 5 uL 23 ng/ul pMM65
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 11 |
| 1 | 65 | 5 |
| 1 | 72 | 15 |
| 30 | 98 | 1 |
| 72 | 15 | |
| 1 | 72 | 300 |
| 1 | 4 | inf |
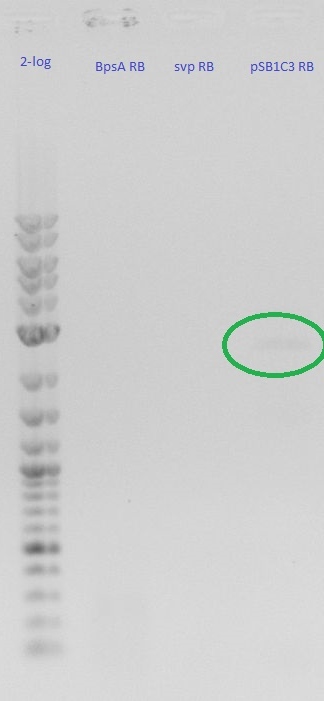
Agarose Gel
- pSB1C3 backbone
- weak band on gel < 50 ng/ 17 ul
- Troubleshooting
- two step PCR -> try annealing step with 66 °C annealing temp (NEB Tm Calculator Phusion)
- circular template
- template sequence?
- bpsA
- cells not lysed? DNA damaged (nitrogen)?
- svp
#2 PCR pSB1C3 RB09/10
- 8 ul water
- 10 ul Phusion Flash HF MM
- je 0,5 ul Primer 100 uM
- 1 ul pSB1C3 1:5
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 62 | 5 | |
| 72 | 45 | |
| 1 | 72 | 180 |
| 1 | 4 | inf |
PCR svp RB07/08 from pMM65 23 ng/ ul circular
- 8 ul water
- 10 ul Phusion Flash HF MM
- je 0,5 ul Primer 100 uM
- 1 ul pMM65 1:10
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 15 |
| 30 | 98 | 1 |
| 62 | 5 | |
| 72 | 15 | |
| 1 | 72 | 300 |
| 1 | 4 | inf |
PCR bpsA RB05/06 from S. lavendulae
- Streptomyces was washed 3x in H20 dest; 2 "colonies" in 1 uL
- 8 ul water
- 10 ul Phusion Flash HF MM
- je 0,5 ul Primer 100 uM
- 1 ul Streptomyces culture
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 180 |
| 30 | 98 | 1 |
| 62 | 5 | |
| 72 | 60 | |
| 1 | 72 | 300 |
| 1 | 4 | inf |
0,8 % Agarose Gel
- pSB1C3 worked well with 20-50 ng/ ul
- BpsA
- annealing temp still too high?
- cell lysate destroys master mix? -> nitrogen/ boil before PCR, use liquid phase
- cells not lysed? vortex with glas beads
#3 desperation PCR
| PCR | water | Phusion Flash HF MM | Primer 100 uM | DMSO | template | template |
|---|---|---|---|---|---|---|
| Ia | 6 | 10 | RB05, RB06 à 0,5 ul | 1 | 2 | S. lav, medium 65, nitrogen shock, glass beads vortex, 98 °C with water
bidest; liquid phase |
| Ib | 6 | 10 | RB05, RB06 à 0,5 ul | 0 | 3 | S. lav, medium 65, nitrogen shock, glass beads vortex, 98 °C with water
bidest; liquid phase |
| II | 6 | 10 | RB05, RB06 à 0,5 ul | 0 | 3 | S. lav, medium 65, glass beads vortex, 98 °C with water bidest; liquid phase |
| III | 6 | 10 | RB05, RB06 à 0,5 ul | 0 | 3 | S. lav, medium 65, nitrogen shock, 98 °C with water bidest; liquid phase |
| IVa | 6 | 10 | RB05, RB06 à 0,5 ul | 1 | 2 | S. lav, medium 65, 98 °C with water bidest; liquid phase |
| IVb | 6 | 10 | RB05, RB06 à 0,5 ul | 0 | 3 | S. lav, medium 65, 98 °C with water bidest; liquid phase |
| V | 6 | 10 | RB05, RB06 à 0,5 ul | 0 | 3 | S. lav, medium 65, nitrogen shock, glass beads vortex; pellet |
| VI | 6 | 10 | RB05, RB06 à 0,5 ul | 0 | 3 | S. lav, YEME medium, nitrogen shock, glass beads vortex; pellet/ liquid
phase |
| VII | 6 | 10 | RB05, RB06 à 0,5 ul | 0 | 3 | S. lav, YEME medium, nitrogen shock, 98 °C with water bidest; liquid phase |
| VIII | 7 | 10 | RB07, RB08 à 0,5 ul | 1 | 1 | pMM65 2,3 ng/ ul, trace RB05 |
| IX | 6 | 10 | RB07, RB08 à 0,5 ul | 0 | 1 | pMM65 2,3 ng/ ul, should be 8 ul water |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 120 |
| 2 | 98 | 1 |
| 40 | 5 | |
| 72 | 70 | |
| 2 | 98 | 1 |
| 45 | 5 | |
| 72 | 70 | |
| 5 | 98 | 1 |
| 50 | 5 | |
| 72 | 70 | |
| 5 | 98 | 1 |
| 55 | 5 | |
| 72 | 70 | |
| 15 | 98 | 1 |
| 65 | 5 | |
| 72 | 70 | |
| 1 | 72 | 300 |
| 1 | 4 | inf |
Desperation Gel
- band around 200 bp? unspecific product due to PCR conditions, but then why no product?
- VI glowing pocket -> sucrose in YEME medium? -> PCR purification and new gel -> still nothing
Phusion Flash HF has proofreading activity; so mutations may stop elongation -> try intron iTaq and design new
primers without mutations.
- PCR svp from NI7/8 PCR product and pMM65, respectively; svp with NI7/8 as control; intron iTaq 2x Master mix.
| PCR | iTaq 2x Master Mix | template | Primer 10 uM | water |
|---|---|---|---|---|
| A | 10 ul | pMM65 2 ul 2,3 ng/ ul | 1 ul RB07 and RB08 | 6 ul |
| B | 10 ul | pMM65 2 ul 2,3 ng/ ul | 2 ul RB07 and RB08 | 4 ul |
| C | 10 ul | svp 2 ul 7,2 ng/ ul | 1 ul RB07 and RB08 | 6 ul |
| D | 10 ul | svp 2 ul 7,2 ng/ ul | 2 ul RB07 and RB08 | 4 ul |
| E | 10 ul | svp 1 ul 7,2 ng/ ul | 1 ul NI07 and NI08 | 8 ul |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 94 | 120 |
| 35 | 94 | 20 |
| 60 | 10 | |
| 70 | 60 | |
| 1 | 72 | 240 |
| 1 | 4 | inf |
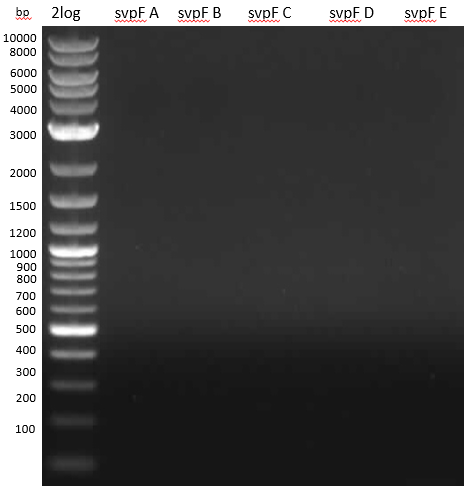
Gel 0,8 % 100 V 60 min
- nothing
- intron iTaq maybe doesn't work so well since control isn't working; or standard protocol is bad.
- run PCR with other Taq Master Mix
- iTaq with Slav colony YEME
| Tube | Master Mix | MM 2x [ul] | Primer 100 mM [ul] | template | template [ul] | water [ul] |
|---|---|---|---|---|---|---|
| A:svp RB7/8 | Fermentas Taq PCR Master Mix 2x | 25 | 7,5 | svp 7,2 ng/ ul | 10 | 0 |
| B:svp RB7/8 | Fermentas Taq PCR Master Mix 2x | 25 | 7,5 | svp 7,2 ng/ ul | 10 | 0 |
| C:pMM65 RB7/8 | Fermentas Taq PCR Master Mix 2x | 25 | 7,5 | pMM65 2,3 ng/ ul | 10 | 0 |
| D:pMM65 RB7/8 | Fermentas Taq PCR Master Mix 2x | 25 | 7,5 | pMM65 2,3 ng/ ul | 10 | 0 |
| E:pMM65 NI7/8 | Fermentas Taq PCR Master Mix 2x | 25 | 7,5 | pMM65 2,3 ng/ ul | 10 | 0 |
| F:svp RB7/8 | Finnzymes Taq PCR Master Mix 2x | 25 | 7,5 | svp 7,2 ng/ ul | 10 | 0 |
| G:svp RB7/8 | Finnzymes Taq PCR Master Mix 2x | 25 | 7,5 | svp 7,2 ng/ ul | 10 | 0 |
| H:pMM65 RB7/8 | Finnzymes Taq PCR Master Mix 2x | 25 | 7,5 | pMM65 2,3 ng/ ul | 10 | 0 |
| I:pMM65 RB7/8 | Finnzymes Taq PCR Master Mix 2x | 25 | 7,5 | pMM65 2,3 ng/ ul | 10 | 0 |
| J:pMM65 NI7/8 | Finnzymes Taq PCR Master Mix 2x | 25 | 7,5 | pMM65 2,3 ng/ ul | 10 | 0 |
| K:svp RB7/8 | NEB OneTaq Quick Load PCR Master Mix 2x | 25 | 7,5 | svp 7,2 ng/ ul | 10 | 0 |
| L:svp RB7/8 | NEB OneTaq Quick Load PCR Master Mix 2x | 25 | 7,5 | svp 7,2 ng/ ul | 10 | 0 |
| M:pMM65 RB7/8 | NEB OneTaq Quick Load PCR Master Mix 2x | 25 | 7,5 | pMM65 2,3 ng/ ul | 10 | 0 |
| N:pMM65 RB7/8 | NEB OneTaq Quick Load PCR Master Mix 2x | 25 | 7,5 | pMM65 2,3 ng/ ul | 10 | 0 |
| O:pMM65 NI7/8 | NEB OneTaq Quick Load PCR Master Mix 2x | 25 | 7,5 | pMM65 2,3 ng/ ul | 10 | 0 |
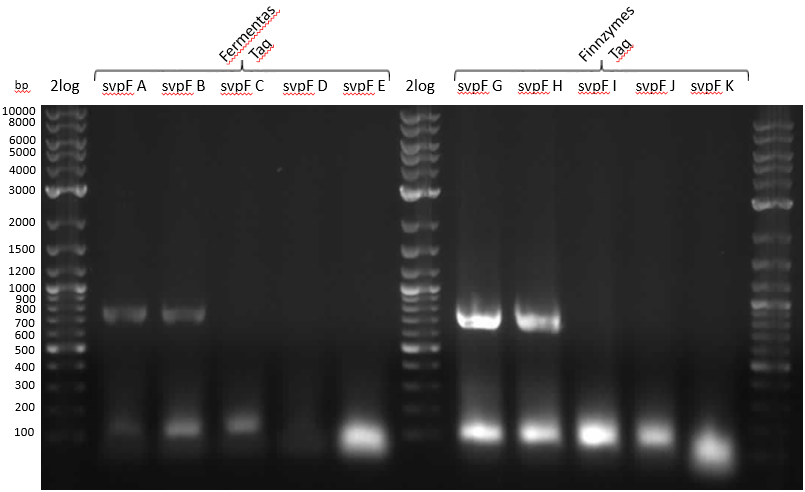
- gel analysis
- svp worked well as template for RB07/08, but lots of wrong product around 100-150 bp. Maybe primers bind to each other, folding results in other preferred sequence or gibson overlap has big affinity though alignments didn't show that
- pMM65 amplification only with NEB OneTaq
- no product with NI07/08 primers...because they are for TE-domain of BpsA. Ni09/10 would have been the right choice
- for further runs, use OneTaq and Fermentas Taq
- was svp amplified or is it just template?
- Again glowing pocket in Slav VI
- huge amount of primers was used in al reactions
PCR with Phusion Flash HF from PCR products (yesterday with Taq-polymerases)
- 10 ul MM; Primer je 2 ul unv.; water 4 ul; template 2 ul
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 55 | 5 | |
| 72 | 30 | |
| 1 | 72 | 300 |
| 1 | 4 | inf |
gel 9 ul 1x loading buffer, 1 ul pcr product
- gel analysis
- no significant amplification of svp with Phusion Flash
- Taq PCR amplified template
- run different protocol
- PCR Phusion flash taq-amplified svp 2
- 10 ul MM, 1 ul primer, 7 ul water, 1 ul water
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 30 | |
| 1 | 72 | 180 |
| 1 | 4 | inf |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72 | 35 | |
| 1 | 72 | 180 |
| 1 | 4 | inf |
gel with 9 ul 1x loading buffer and 1 ul pcr product
- gel analysis
- 3-step worked better than 2-step
- primers might prime each other or the fragment
- it's not just too much primer because lane 4/5 are a lot brighter than 6/7 -> the 120 bp-band is an
amplification product. -> order S. verticillus and published primers from Takahashi and Sanchez -> order published Primers for sfp, entD (Lambalot), bpsA (Takahashi) -> order stuff for DNA isolation of Streptomyces
 "
"