Team:Heidelberg/Templates/Indigoidine week13
From 2013.igem.org
(Created page with " ===Indigoidine production Konrad (Konrad)=== [[File:20130714-f7-delRest.png|400px|thumb|right|PCR for amplification of pSB1C3: ('''left''') first analytical gel; ('''right'''...") |
|||
| Line 2: | Line 2: | ||
===Indigoidine production Konrad (Konrad)=== | ===Indigoidine production Konrad (Konrad)=== | ||
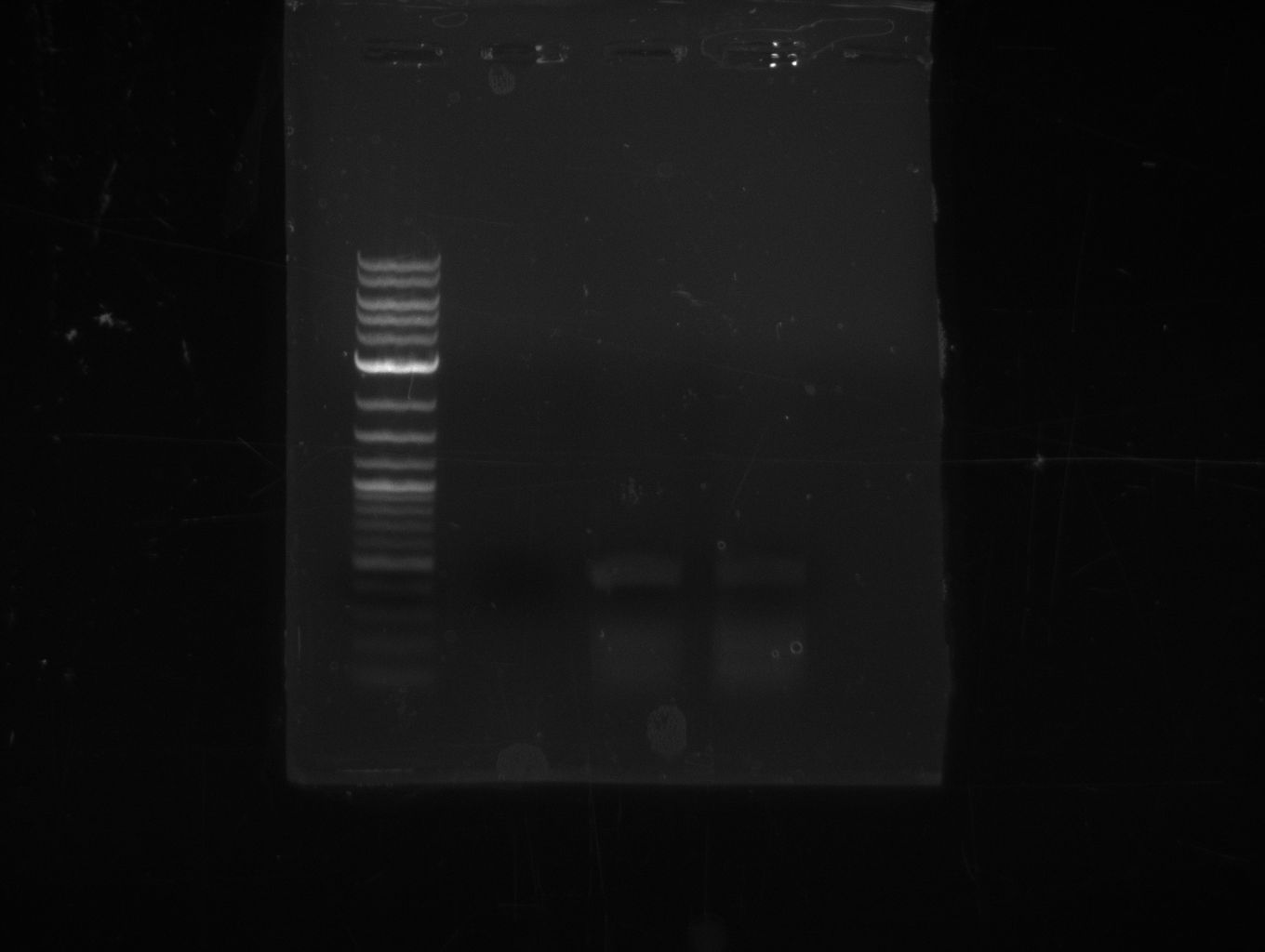
| - | [[File: | + | [[File:Heidelberg_20130714-f7-delRest.png|400px|thumb|right|PCR for amplification of pSB1C3: ('''left''') first analytical |
gel; ('''right''') second analytical gel. f7 was twice eluted after gel extraction, the second run was 30 min in 15 | gel; ('''right''') second analytical gel. f7 was twice eluted after gel extraction, the second run was 30 min in 15 | ||
| - | µl H<sub>2</sub>O at 50 | + | µl H<sub>2</sub>O at 50 °Ca before centrifugation; run at 100 V, 0.8 % gel (TAE);]] |
* gel extraction of yesterdays fragment f7:pSB1C3 | * gel extraction of yesterdays fragment f7:pSB1C3 | ||
* analytical gel (small, half size): (M: 3 µl NEB log2, 2 µl f7 + 2 µl 1x loading dye)<br/>==> [f7] = 3 ng/µl | * analytical gel (small, half size): (M: 3 µl NEB log2, 2 µl f7 + 2 µl 1x loading dye)<br/>==> [f7] = 3 ng/µl | ||
| Line 20: | Line 20: | ||
enrichment of desired fusion product with fw primer of fragment 1 and rv primer of fragment 2. | enrichment of desired fusion product with fw primer of fragment 1 and rv primer of fragment 2. | ||
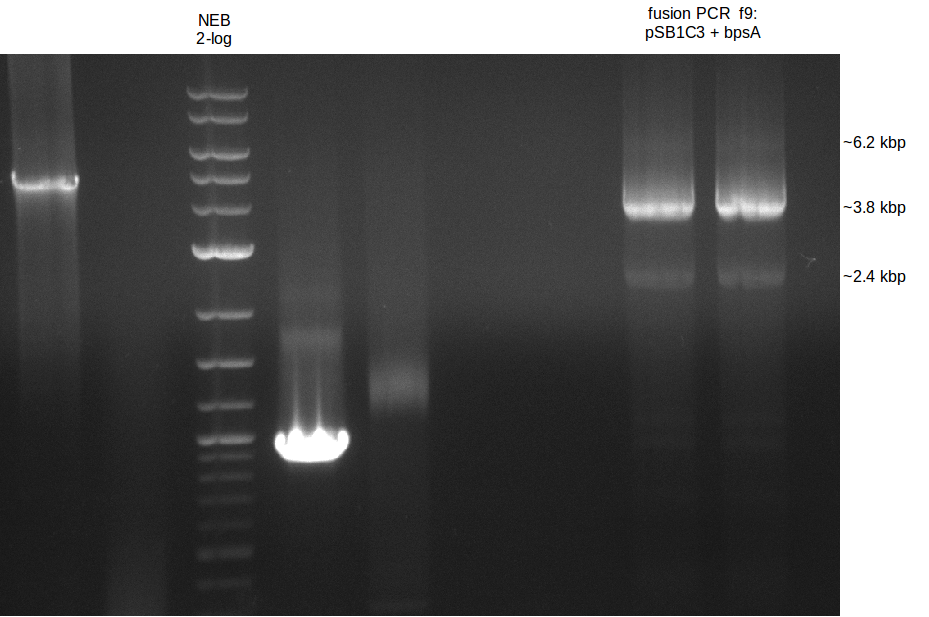
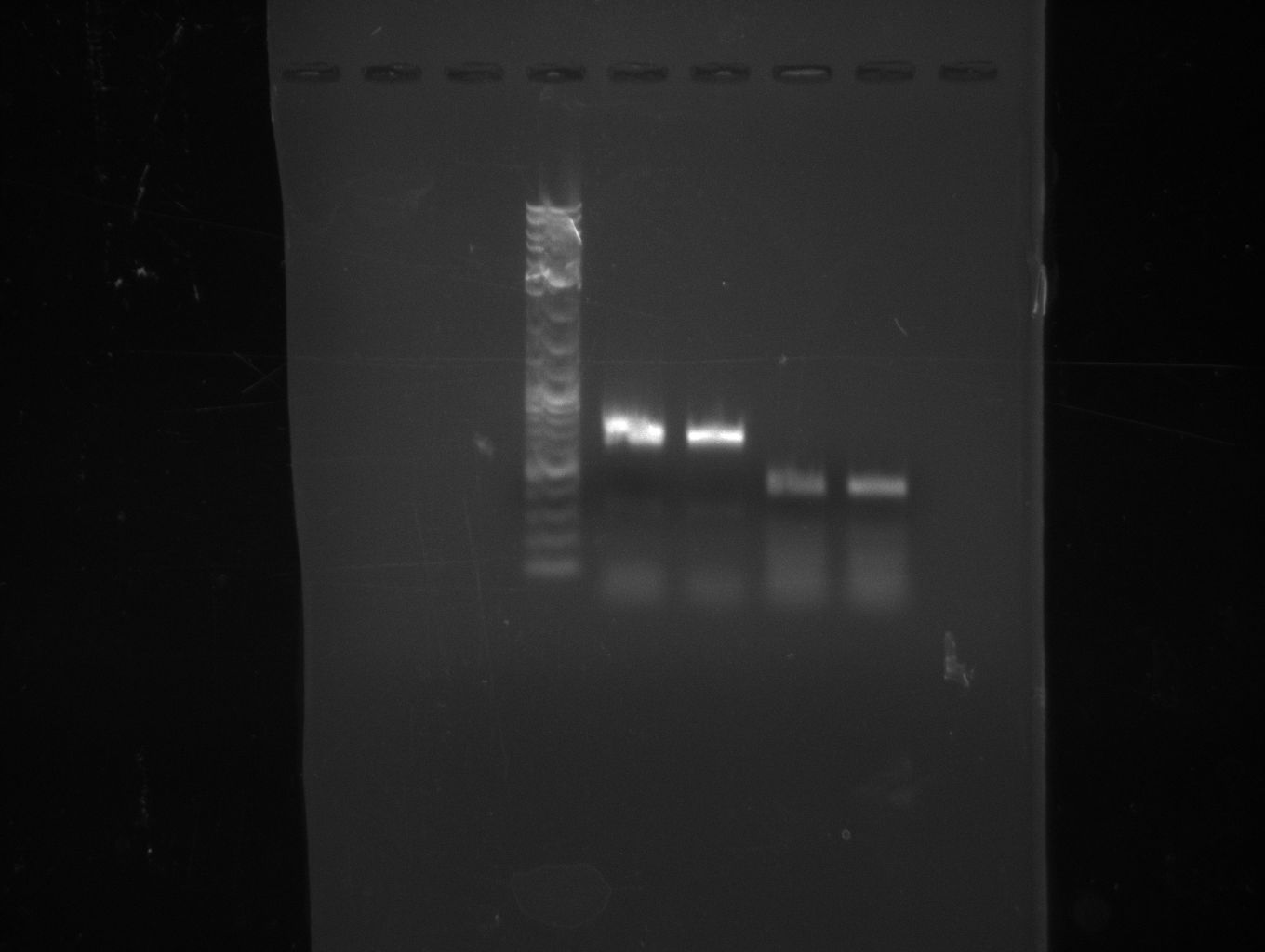
| - | [[File: | + | [[File:Heidelberg_090714-f9.png|400px|thumb|right|PCA in order to gain bpsA and pSB1C3 as one fragment fused together. The |
wanted fragment size is ~ 6.2 kbp.]] | wanted fragment size is ~ 6.2 kbp.]] | ||
| Line 64: | Line 64: | ||
==== Preparation for electroporation ==== | ==== Preparation for electroporation ==== | ||
| - | [[File: | + | [[File:Heidelberg_20130714-GA.png|200px|thumb|right|analytical gel of purified Gibson assembly and delRest;run at 100 V, 0.8 % |
gel (TAE);]] | gel (TAE);]] | ||
| Line 73: | Line 73: | ||
==== Validation of Gibson assembly ==== | ==== Validation of Gibson assembly ==== | ||
| - | [[File: | + | [[File:Heidelberg_20130715-GA.png|400px|thumb|right|Gibson assembly. The wanted fragment size is ~ 7.0 kbp.;run at 100 V, 0.8 |
% gel (TAE);]] | % gel (TAE);]] | ||
| Line 80: | Line 80: | ||
==== fusion PCR (bpsA+svp) ==== | ==== fusion PCR (bpsA+svp) ==== | ||
| - | [[File: | + | [[File:Heidelberg_20130715-PCA.png|200px|thumb|right|PCA in order to gain bpsA and pSB1C3 as one fragment fused together. The |
wanted fragment size is ~ 6.2 kbp.;run at 135 V, 0.8 % gel (TAE);]] | wanted fragment size is ~ 6.2 kbp.;run at 135 V, 0.8 % gel (TAE);]] | ||
| Line 108: | Line 108: | ||
* experimental CPEC assembly with left mix from day before | * experimental CPEC assembly with left mix from day before | ||
| - | [[File: | + | [[File:Heidelberg_20130719-CPEC.png|100px|thumb|right|Analytical gel for verification of CPE success.]] |
| - | File: | + | File:Heidelberg_20130719-CPEC.png |
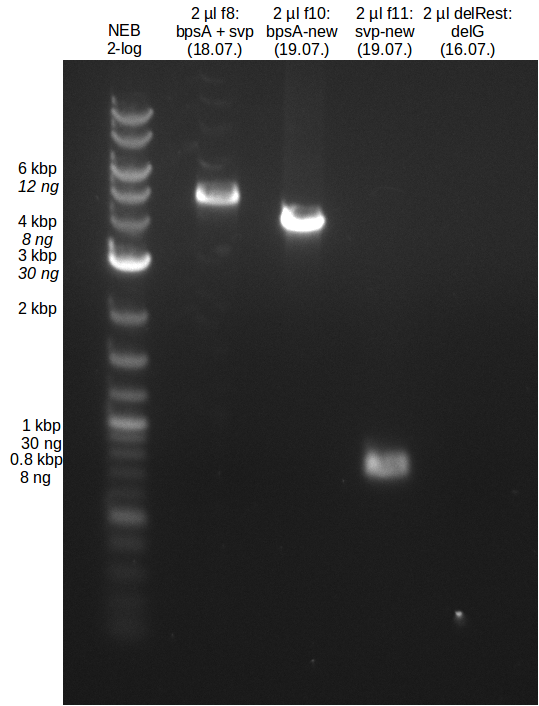
* PCR of bpsA and svp with new Primers | * PCR of bpsA and svp with new Primers | ||
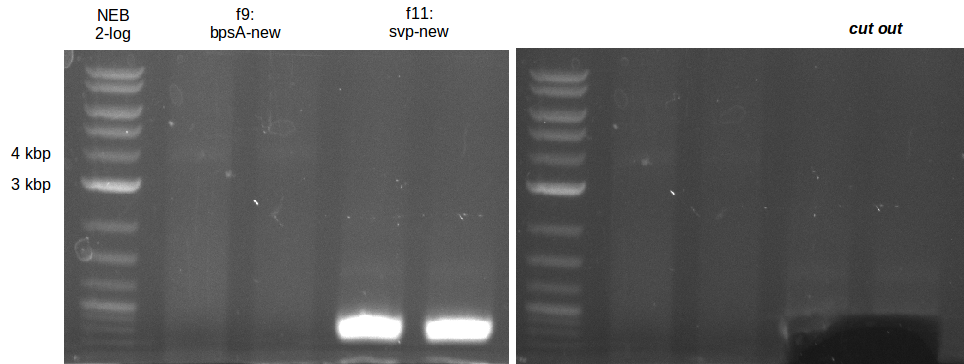
| - | [[File: | + | [[File:Heidelberg_20130719-svp_new.png|200px|thumb|right|PCR amplification of svp with new Primers.]] |
| - | [[File: | + | [[File:Heidelberg_20130719-bpsA_new.png|100px|thumb|right|PCR amplification of bpsA with new Primers.]] |
* gel extraction of new bpsA and svp fragments | * gel extraction of new bpsA and svp fragments | ||
| - | [[File: | + | [[File:Heidelberg_20130719-GE-f8_10_11-delRest.png|100px|thumb|right|Analytical gel bpsA and svp gel extraction.]] |
* Gibson Assembly | * Gibson Assembly | ||
* CPEC | * CPEC | ||
| - | [[File: | + | [[File:Heidelberg_20130719-GA-CPEC.png|100px|thumb|right|Analytical gel for verification of Gibson assembly and CPEC |
success.]] | success.]] | ||
| Line 123: | Line 123: | ||
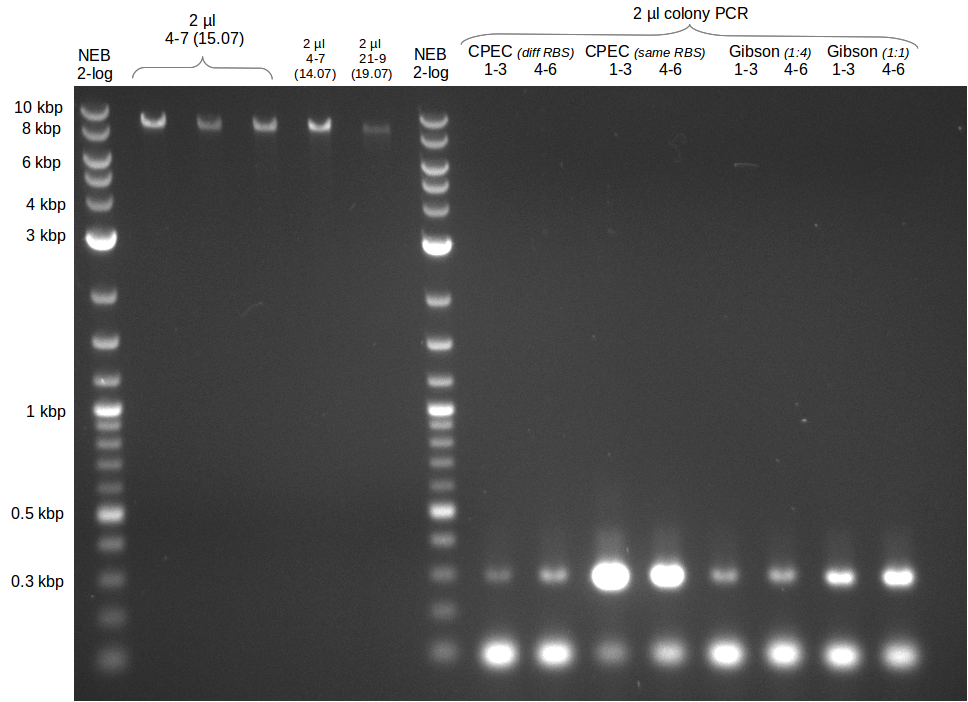
* Colony PCR of electroporation with GA | * Colony PCR of electroporation with GA | ||
| - | [[File: | + | [[File:Heidelberg_20130720-CP_GA_electroporation.png|500px|thumb|right|Colony PCR for pKH1 validation. The T-TE domain of bpsA |
was used as amplicon (size: 1.1 kbp) as well as svp-part of backbone (similar size of 1.1 kbp). Every lane | was used as amplicon (size: 1.1 kbp) as well as svp-part of backbone (similar size of 1.1 kbp). Every lane | ||
| Line 129: | Line 129: | ||
represents one PCR reaction with 1 picked colony each. Except for colony 1 only black colonies were picked.]] | represents one PCR reaction with 1 picked colony each. Except for colony 1 only black colonies were picked.]] | ||
* Colony PCR of trafos of day before | * Colony PCR of trafos of day before | ||
| - | [[File: | + | [[File:Heidelberg_20130721-delRest4-7_21-9_colony.png|500px|thumb|right|Colony PCR for pKH1 und pKH2 validation. The T domain |
of bpsA was used as amplicon (size: 300 bp). Every lane represents one PCR reaction with 3 picked colony each.]] | of bpsA was used as amplicon (size: 300 bp). Every lane represents one PCR reaction with 3 picked colony each.]] | ||
| Line 195: | Line 195: | ||
<gallery widths="400px" heights="300px"> | <gallery widths="400px" heights="300px"> | ||
| - | + | File:Heidelberg_20130724_pMM64-RB-3set.jpg|bpsA; entD-l; svpV-l; svpF-l; entD-s; svpV-s | |
| - | + | File:Heidelberg_20130724_pMM64-RB-3set_cut.jpg|bpsA; entD-l; svpV-l; svpF-l; entD-s; svpV-s | |
</gallery> | </gallery> | ||
| Line 225: | Line 225: | ||
<gallery widths="400px" heights="300px"> | <gallery widths="400px" heights="300px"> | ||
| - | + | File:Heidelberg_20130726_PPTasenhasen.jpg|1 ug 2-log; entDlong; svpVlong; svpFlong | |
| - | + | File:Heidelberg_20130726_PPThasen_cut.jpg|1 ug 2-log; entDlong; svpVlong; svpFlong | |
</gallery> | </gallery> | ||
| Line 253: | Line 253: | ||
<gallery widths="400px" heights="300px"> | <gallery widths="400px" heights="300px"> | ||
| - | + | File:Heidelberg_20130726_svpV.jpg|2-log; RB13/14 65°C; RB39/40 62 °C; RB 39/40 57 °C | |
</gallery> | </gallery> | ||
| Line 280: | Line 280: | ||
<gallery widths="400px" heights="300px"> | <gallery widths="400px" heights="300px"> | ||
| - | + | File:Heidelberg_20130728rb_svpverttaka.jpg|2-log; Hanna; svpV RB13/14 | |
| - | + | File:Heidelberg_20130728rb_svpverttaka_cut.jpg|2-log; Hanna; svpV RB13/14 | |
</gallery> | </gallery> | ||
| Line 313: | Line 313: | ||
<gallery widths="400px" heights="300px"> | <gallery widths="400px" heights="300px"> | ||
| - | + | File:Heidelberg_20130728_resvpVlong.jpg|2-log; from taka; from RB39/40 | |
| - | + | File:Heidelberg_20130728_resvpVlong_cut.jpg|2-log; from taka; from RB39/40 | |
</gallery> | </gallery> | ||
Latest revision as of 00:47, 5 October 2013
Contents |
Indigoidine production Konrad (Konrad)
- gel extraction of yesterdays fragment f7:pSB1C3
- analytical gel (small, half size): (M: 3 µl NEB log2, 2 µl f7 + 2 µl 1x loading dye)
==> [f7] = 3 ng/µl - 2nd analytical gel (small, half size): (M: 3 µl NEB log2, (2 µl f1:4.07., 2 µl f7:14.07., 2 µl f7:14.07.-2nd
eluation, 2 µl delRest-f6-9) + 8 µl 1x loading dye)
==> [f1:4.07.] = 18 ng/µl, [f7:14.07.] = 5 ng/µl,
[f7:14.07.-2nd] = 2 ng/µl,[delRest-f] = 5 ng/µl
fusion PCR (bpsA+svp)
based on PCA protocol: Used two main steps, first elongation of fragments (f1;f7) without any primers, than
enrichment of desired fusion product with fw primer of fragment 1 and rv primer of fragment 2.
Step 1:
- 10 µl Phusion Flash MM
- 6.3 µl template (f7:pSB1C3)
- 3.7 µl template (f1:bpsA from 4.07.)
Step 2:
- 20.0 µl H2O
- 25 µl Phusion Flash MM
- 4.0 µl template (reaction from step 1)
- 2x 0.5 µl primer (NI:06; NI:09)
| Step | Cycles | temperature [°C] | Time [min] |
|---|---|---|---|
| 1 | 1 | 98 | 0:10 |
| 5 | 98 | 0:01 | |
| 52 | 0:30 | ||
| 72 | 0:20 | ||
| 1 | 72 | 5:00 | |
| 1 | 12 | inf | |
| use only 4.0 µl of mixture for 2nd step | |||
| 2 | 1 | 98 | 0:10 |
| 30 | 98 | 0:01 | |
| 72 | 1:10 | ||
| 1 | 72 | 5:00 | |
| 1 | 12 | inf | |
Preparation for electroporation
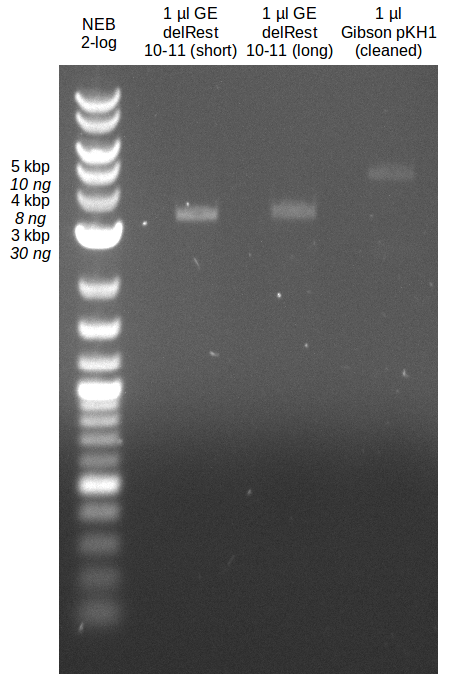
- Gibsonassembly purified with Nucleotide Removal Kit
- analytical gel (small, half size): (M: 2.5 µl NEB log2)
==> [delRest(short)] = 6-7 ng/µl, [delRest(long)] = 4
ng/µl, [GA] = 3 ng/µl
Validation of Gibson assembly
- put 10 µl on gel.
- electroporation with left 4 µl
fusion PCR (bpsA+svp)
Redo 2nd step with 1st step result of the 13th of July
Step 2:
- 23.5 µl H2O
- 25 µl Phusion Flash MM
- 0.5 µl template (reaction from step 1)
- 2x 0.5 µl primer (NI:06; NI:09)
| Step | Cycles | temperature [°C] | Time [min] |
|---|---|---|---|
| 2 | 1 | 98 | 0:10 |
| 30 | 98 | 0:01 | |
| 72 | 1:10 | ||
| 1 | 72 | 5:00 | |
| 1 | 12 | inf |
soil sample preparation
- experimental CPEC assembly with left mix from day before
File:Heidelberg_20130719-CPEC.png
- PCR of bpsA and svp with new Primers
- gel extraction of new bpsA and svp fragments
- Gibson Assembly
- CPEC
- Trafo
- Colony PCR of electroporation with GA
- Colony PCR of trafos of day before
- Heat-shock competent Rosetta
- Minipreparation of 2 ml ON culture of one picked black colony of GA plate (electroporation)
Ralf
PCR PPTases #1 Phusion Flash HF 50 ul (long) 20 ul (short); colony pick/0.2 ul pcr template; primer 2/5 ul 10 uM
- MG1655 pick RB33/34 and RB 41/42
- S. vert pick liquid RB29/30 and RB 39/40
- pMM65 pcr RB25/26
biorad T100
| cycles | temperature °C | time seconds |
|---|---|---|
| 1 | 98 | 180/10 |
| 12 | 98 | 1 |
| td 68 -0.5 | 5 | |
| 72 | 20 | |
| 18 | 98 | 1 |
| 65 | 5 | |
| 72 | 20 | |
| 1 | 72 | 180 |
| 1 | 12 | - |
PCR bpsA64 Phusion Flash HF 50 ul (long); 0.2 ul pcr template; primer 5 ul 10 uM
- pMM64 Fussenegger miniprep RB23/24
biometra
| cycles | temperature °C | time seconds |
|---|---|---|
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| td 58 -0.5 | 5 | |
| 72 | 70 | |
| 18 | 98 | 1 |
| 65 | 5 | |
| 72 | 70 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
PCR PPTases #3 Phusion Flash HF 50 ul (25 MM; 5/5 primer; 14.5 water; 0.5 template)
- 1: entD from PPTases #1 gel ex w/ RB33/34 long
- 2: svpV from ??? w/ RB29/30 long
- 3: svpF from PPTases #1 gel ex w/ RB25/26 long
| cycles | temperature °C | time seconds |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 20 | |
| 1 | 72 | 180 |
| 1 | 12 | - |
analysis
- entD and svpF -> gel extraction
- svpV is too small -> wrong product. new colony PCR with short primers
Konrad PCR svpV #3
| cycles | temperature °C | time seconds |
|---|---|---|
| 1 | 98 | 180 |
| 30 | 98 | 1 |
| 3grad. 65-62-57 | 5 | |
| 72 | 20 | |
| 1 | 72 | 180 |
| 1 | 10 | - |
analysis
- hmm
PCR svpV #4 20 ul Phusion Flash HF (10 MM; 9.4 water; 0.2 ul primer 100 uM; 0.2 cell pellet) conditions as in Jul 12th; RB13/14 biorad T100
| cycles | temperature °C | time seconds |
|---|---|---|
| 1 | 98 | 120 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 20 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
rePCR svpV #5 from #4 50 ul (25 MM; 14 water; 0.5 primer 100 uM; 0.2 template)
- 1: RB29/30 from short RB13/14
- 2: RB29/30 from short RB 39/40
biorad T100
| cycles | temperature °C | time seconds |
|---|---|---|
| 1 | 98 | 10 |
| 10 | 98 | 1 |
| td 65 -0.5 | 5 | |
| 72 | 20 | |
| 25 | 98 | 1 |
| 65 | 5 | |
| 72 | 20 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
-> gel extraction; CPEG assembly
| plasmid name | backbone | cutting site | indigoidine-Synthetase | cutting site | PPTase | cutting site | primers |
|---|---|---|---|---|---|---|---|
| pRB3 | pSB1C3-BBa_B0034 | KpnI | P.lum indC | BamHI | Bsub sfp | NheI | RB21/22-27/28-35/36 |
| pRB4 | pSB1C3-BBa_B0034 | KpnI | P.lum indC | BamHI | Svert svp | NheI | RB21/22-27/28-29/30 |
| pRB5 | pSB1C3-BBa_B0034 | KpnI | P.lum indC | BamHI | Svert svp65 | NheI | RB21/22-27/28-25/26 |
| pRB6 | pSB1C3-BBa_B0034 | KpnI | P.lum indC | BamHI | Eco entD | NheI | RB21/22-27/28-33/34 |
| pRB7 | pSB1C3-BBa_B0034 | KpnI | S.lav bpsA64 | BamHI | Bsub sfp | NheI | RB21/22-23/24-35/36 |
| pRB8 | pSB1C3-BBa_B0034 | KpnI | S.lav bpsA64 | BamHI | Svert svp | NheI | RB21/22-23/24-29/30 |
| pRB9 | pSB1C3-BBa_B0034 | KpnI | S.lav bpsA64 | BamHI | Svert svp65 | NheI | RB21/22-23/24-25/26 |
| pRB10 | pSB1C3-BBa_B0034 | KpnI | S.lav bpsA64 | BamHI | Eco entD | NheI | RB21/22-23/24-33/34 |
CPEG assembly 20 ul Phusion Flash HF (10 MM; 2 pSB1C3; 1.5 PPTase; 6 ind synthetase)
biometra
| cycles | temperature °C | time seconds |
|---|---|---|
| 1 | 98 | 10 |
| 10 | 98 | 1 |
| 55 | 5 | |
| 72 | 70 | |
| 1 | 72 | 180 |
| 1 | 12 | - |
Transformation 10 pm; 80 ul +/- 20 ul; 5 ul CPEG pcr product
- medium LB: FG 2013Jul19
- plates LB+Cm: FG 2013Jul22
- TOP10 from stock -80 °C
 "
"