Team:Shenzhen BGIC ATCG/Project
From 2013.igem.org
(→Actor - signal peptides) |
(→Targeting Peptide) |
||
| Line 85: | Line 85: | ||
The ZRC1 gene encodes a multicopy suppressor of zinc toxicity in Saccharomyces cerevisiae; however, previously it was reported that the expression of ZRC1 was induced when the intracellular zinc level was decreased. The COT1 and ZRC1 genes of Saccharomyces cerevisiae are structurally related dosage-dependent suppressors of metal toxicity. COT1 confers increased tolerance to high levels of cobalt; ZRC1 confers increased tolerance. 【Interactions between gene products involved in divalent cation transport in Saccharomyces cerevisiae.】Zrc1 has six putative transmembrane domains, and Zrc1-GFP fusion protein was localized to the vacuolar membrane. Zrc1 function as a mechanism to maintain the zinc homeostasis in yeast. 【The Zrc1 is involved in zinc transport system between vacuole and cytosol in Saccharomyces cerevisiae.】 | The ZRC1 gene encodes a multicopy suppressor of zinc toxicity in Saccharomyces cerevisiae; however, previously it was reported that the expression of ZRC1 was induced when the intracellular zinc level was decreased. The COT1 and ZRC1 genes of Saccharomyces cerevisiae are structurally related dosage-dependent suppressors of metal toxicity. COT1 confers increased tolerance to high levels of cobalt; ZRC1 confers increased tolerance. 【Interactions between gene products involved in divalent cation transport in Saccharomyces cerevisiae.】Zrc1 has six putative transmembrane domains, and Zrc1-GFP fusion protein was localized to the vacuolar membrane. Zrc1 function as a mechanism to maintain the zinc homeostasis in yeast. 【The Zrc1 is involved in zinc transport system between vacuole and cytosol in Saccharomyces cerevisiae.】 | ||
| - | === | + | === Make-up Artists - introns === |
| + | SRC1 intron has two 5’ splicing site whose efficiency is regulated by protein hub1. Originally, the existence of the intron would produce two mature mRNA in proportion. After engineering, the existence of protein hub1 regulates to the preservation of intron precisely, which would make the following mRNA be expressed normally or not. As for the silencing of wild type gene HUB1, we choose CRISPRi that is comparably easy to use reversibly. | ||
| + | |||
| + | ***Scr1 and Hub1 | ||
| + | |||
| + | Alternative splicing substantially increases the gene product diversity and is a major source of cell type differentiation. A good example is the alternative splicing of Saccharomyces cerevisiae SRC1 pre-mRNA, which is promoted by the conserved ubiquitin-like protein Hub1. It can function through binding non-covalently to a conserved element termed HIND in the spliceosomal protein Snu66. Such binding makes the splicesome target sites change and moderately alters spliceosomal interactions. | ||
| + | Hub1 is a ubiquitin-like modifier (UBL) that covalent modify the proteins. Interest enough, it harbors several different to other UBLs in which it possesses a C-terminal double tyrosine motif while others having a GG motif. The Snu66, a tri-snRNP in yeast spliceosome, possesses with two N-terminal HINDs (Hub1-INteraction Domain). The Hub1–HIND interaction comprises a strong salt bridge accompanied by several hydrophobic contacts and high affinity. Such binding modifies the spliceosome rather than modulating the properties of an individual binding partner. Hub1-controlled splicing occurs universally in eukaryotes. SR proteins and hnRNPs involved in spliceosome targeting do not seem to exist in S. Cerevisiae , and thus the Hub1-dependent mechanism may be evolutionarily older. | ||
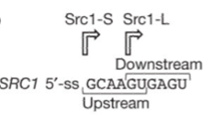
| + | Scr1 is a protein in yeast having alternative splicing sites in its intron. The characteristic differential Hub1 dependence of SRC1 alternative splicing requires the tandem arrangement of overlapping 5’ splice sites. The Hub1 binding spliceosome can splice the intron from both downstream 5’ sites as well as the upstream 5’ sites with preference to the former one. | ||
| + | |||
| + | [[File:Figure3._The_SRC_intron_with_two_splice_sites.png]] | ||
| + | |||
| + | Showed in Figure.1 When cutting in the upstream splice sites, the exons flanking around it would be translated as a fusion protein Scr-S. However, when cutting at the down stream one, the left 4bp in intron would result in a frameshift, thus only the forward exon can express named Scr-L. | ||
| + | |||
| + | ***dCas9 CRISPR interface system: | ||
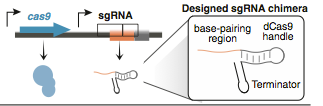
| + | CRISPR shorts for Clustered Regularly Interspaced Palindromaic Repeats system, which can be targeted to DNA using RNA, enabling genetic editing of any region of the genome in many organisms.[Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease .] In the type II CRISPR/Cas system, a ribonucleoprotein complex formed from a single protein (Cas9), a crRNA, and a trans-acting CRISPR RNA (tracrRNA) can carry out efficient crRNA-directed recognition and site-specific cleavage of foreign DNA(CRISPR RNA maturation by transencoded small RNA and host factor RNase III.). After mutated the endonuclease domains of the Cas9 protein, it creates a programmable RNA-dependent DNA-binding protein. The sgRNA consists of three domains: a 20 nt complementary region for specific DNA binding, a 42 nt hairpin for Cas9 binding (Cas9 handle), and a 40 nt transcription terminator derived from S. Pyogenes. After translation, the Cas9 binds to sgRNA to form a protein-RNA complex, which can recognize target sites in the genome sequence and bind to it. Then, it could block RNA polymerase and transciption elongation. | ||
| + | |||
| + | [[File:Figure4._The_structure_of_sgRNA.png ]] | ||
=== Degradation Peptide === | === Degradation Peptide === | ||
=== Editor - the promoters === | === Editor - the promoters === | ||
Revision as of 07:52, 7 September 2013
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Modeling | Notebook | Safety | Attributions |
|---|
Contents |
Overall project
This year, our project named "Cell Magic" we engineered the budding yeast in time level as well as space level. The "Cell Magic", like a movie,can be divided into two parts: the actor related and the time regulation related one. In the first part, our actor, signal peptides, were all made up with colorful clothes by the fluorescent proteins. Furthermore, the intron and degradation biobricks as make-up artists can decide how the clothes be matched. Through these steps, our actors may wearing in green, yellow or even mixed, appear in their specific sub-locations of yeast cell. With respect to the time level, we took advantages of the natural cell cycle in budding yeast to improve the time process to be more suitable for our movie.In general, the movie director, the promoters from five cycline gene, can decide the expression of downstream gene in G1 and G2 phase phase, respectively. Also, freezer Sic1 help our movie stay a longer time in the G1 phase which is important for the actor performing their stories. And the last tools we utilized is the microfluidics, through which numerous cells can project our "Cell Magic", thus ensuring our observation by naked eyes.
Project Details
Actor - signal peptides
A target peptide is a short (3-70 amino acids long) peptide chain that directs the transport of a protein to a specific region in cell, including nucleus, mitochondria, endoplasmic reticulums (ERs), chloroplasts, apoplasts, peroxisomes and plasma membrane. Targeting peptide can exists in both N-terminal, C-terminal and internal sequence of a precursor protein. And after transported, some target peptides are cleaved by signal peptidases. In our project we utilized 19 peptides target to 9 sub-locations in yeast cells, and when combined with fluorescent proteins, such region can be marked by different colors.
- OMM
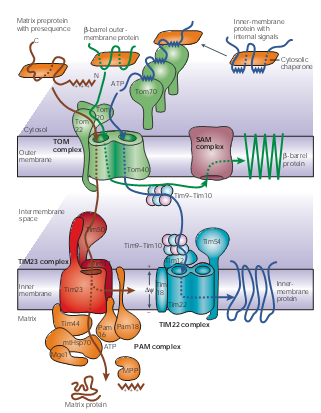
Mitochondria Though it accounts a small ratio in the cell space, mitochondria possess about 10% to 15% proteins encoded by nuclear genes in eukaryotic organisms. These proteins are synthesized in cytosol and then recognized by the membrane receptors of mitochondria. Translocases in the outer and inner membrane of mitochondria mediate the import and intra-mitochondrial sorting of these proteins. ATP is used as an energy source; Chaperones and auxiliary factors assist in folding and assembly of mitochondrial proteins into their native, three-dimensional structures.
Figure 1 | Protein-import pathways for mitochondrial proteins. As shown in figure1, beta-barrel outer-membrane proteins (dark green), precursor proteins (brown) with positively charged amino-terminal presequences and multispanning inner-membrane proteins (blue) with internal targeting signals are recognized by specific receptors of the outer mitochondrial membrane (TOM) translocases Tom20, Tom22 and/or Tom70. The precursor proteins are then translocated through a small Tom proteins of the TOM complex, Tom40 pore, which the TOM complex contains two or three.
- peroxisomes
The import of post-translational matrix protein into peroxisomes depends on either of the two peroxisomal targeting signals (PTS), PTS1 and PTS2. PTS2-driven import is facilitated by a complex in the membrane. Under oleic acid-inducing growth conditions, there is a ternary core complex of approximate 150 kDa in the cytosol, which consists of Fox3p,Pex7p and Pex18p. Fox3p is imported as a dimer, while other two are bind in monomeric forms.
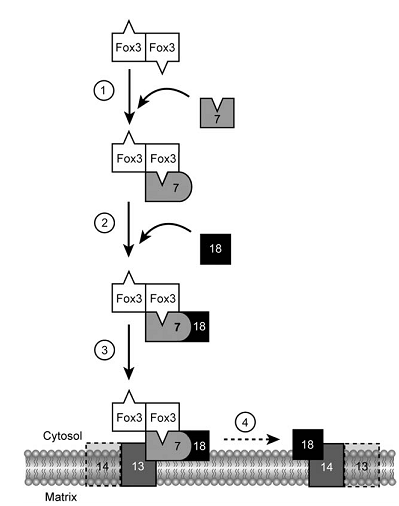 Figure2._Schematical_model_of_the_early_steps_of_PTS2-driven_import.png
Figure2._Schematical_model_of_the_early_steps_of_PTS2-driven_import.png
As study mentioned there are four steps involved in PTS2-driven import. The first step is the recognition of dimerized Fox3p by Pex7p through its PTS2 in the cytosol. In a second step, the Pex7p–Fox3p complex interacts with Pex18p, which targets the PTS2 pre-import complex to the peroxisomal membrane. The third step is the docking process, involving the interaction between Pex7p and the integral membrane protein Pex13p. As a final step of these early steps in the PTS2 import cascade, PTS2 receptor dissociation takes place during or after its assembly into large oligomeric complexes containing Pex14p and Pex13p. Pex18p remains at the peroxisomal membrane in the form of a large-molecular-weight complex in conjunction with Pex14p and/or Pex13p, from where it might be released to the cytosol.
REFERENCE :【Peroxisomal Targeting of PTS2 Pre-Import Complexes in the Yeast Saccharomyces cerevisiae】
- Actin
In yeast, the cortical actin cytoskeleton seems to specify sites of growth of the cell surface. Because the actin-binding protein ABP1p is associated with the cortical cytoskeleton of Saccharomyces cerevisiae, it might be involved in the spatial organization of cell surface growth. ABP1p is localized to the cortical cytoskeleton and its overproduction causes assembly of the cortical actin cytoskeleton at inappropriate sites on the cell surface, resulting in delocalized surface growth. ABP1p is a novel protein with a 50 amino-acid C-terminal domain, which is very similar to the SH3 domain in the non-catalytic region of nonreceptor tyrosine kinases (including those encoded by the proto-oncogenes c-src and c-abl), in phospholipase C gamma and in alpha-spectrin. They also identified an SH3-related motif in the actin-binding tail domain of myosin-I. The identification of SH3 domains in a family of otherwise unrelated proteins that associate with the membrane cytoskeleton indicates that this domain might serve to bring together signal transduction proteins and their targets or regulators, or both, in the membrane cytoskeleton.
REFERENCE: [Homology of a yeast actin-binding protein to signal transduction proteins and myosin-I]
- Nucler
Histones are nuclear proteins package DNA into nucleosomes[Youngson, Robert M. (2006).Collins Dictionary of Human Biology. Glasgow: HarperCollins.], and they are responsible for maintaining the shape and structure of a nucleosome. One chromatin molecule is composed of at least one of each core histones per 100 base pairs of DNA.[The Nucleosome: From Genomic Organization to Genomic Regulation.] There are five families of histones known to date, termed H1/H5, H2A, H2B, H3, and H4.[3] H2A is considered a core histone, along with H2B, H3 and H4. Core formation first occurs through the interaction of two H2A molecules.[Lehninger Principles of Biochemistry.] Then, H2A forms a dimer with H2B; the core molecule is complete when H3-H4 also attaches to form a tetramer.
- vacuolar membrane
The ZRC1 gene encodes a multicopy suppressor of zinc toxicity in Saccharomyces cerevisiae; however, previously it was reported that the expression of ZRC1 was induced when the intracellular zinc level was decreased. The COT1 and ZRC1 genes of Saccharomyces cerevisiae are structurally related dosage-dependent suppressors of metal toxicity. COT1 confers increased tolerance to high levels of cobalt; ZRC1 confers increased tolerance. 【Interactions between gene products involved in divalent cation transport in Saccharomyces cerevisiae.】Zrc1 has six putative transmembrane domains, and Zrc1-GFP fusion protein was localized to the vacuolar membrane. Zrc1 function as a mechanism to maintain the zinc homeostasis in yeast. 【The Zrc1 is involved in zinc transport system between vacuole and cytosol in Saccharomyces cerevisiae.】
Make-up Artists - introns
SRC1 intron has two 5’ splicing site whose efficiency is regulated by protein hub1. Originally, the existence of the intron would produce two mature mRNA in proportion. After engineering, the existence of protein hub1 regulates to the preservation of intron precisely, which would make the following mRNA be expressed normally or not. As for the silencing of wild type gene HUB1, we choose CRISPRi that is comparably easy to use reversibly.
- Scr1 and Hub1
Alternative splicing substantially increases the gene product diversity and is a major source of cell type differentiation. A good example is the alternative splicing of Saccharomyces cerevisiae SRC1 pre-mRNA, which is promoted by the conserved ubiquitin-like protein Hub1. It can function through binding non-covalently to a conserved element termed HIND in the spliceosomal protein Snu66. Such binding makes the splicesome target sites change and moderately alters spliceosomal interactions. Hub1 is a ubiquitin-like modifier (UBL) that covalent modify the proteins. Interest enough, it harbors several different to other UBLs in which it possesses a C-terminal double tyrosine motif while others having a GG motif. The Snu66, a tri-snRNP in yeast spliceosome, possesses with two N-terminal HINDs (Hub1-INteraction Domain). The Hub1–HIND interaction comprises a strong salt bridge accompanied by several hydrophobic contacts and high affinity. Such binding modifies the spliceosome rather than modulating the properties of an individual binding partner. Hub1-controlled splicing occurs universally in eukaryotes. SR proteins and hnRNPs involved in spliceosome targeting do not seem to exist in S. Cerevisiae , and thus the Hub1-dependent mechanism may be evolutionarily older. Scr1 is a protein in yeast having alternative splicing sites in its intron. The characteristic differential Hub1 dependence of SRC1 alternative splicing requires the tandem arrangement of overlapping 5’ splice sites. The Hub1 binding spliceosome can splice the intron from both downstream 5’ sites as well as the upstream 5’ sites with preference to the former one.
Showed in Figure.1 When cutting in the upstream splice sites, the exons flanking around it would be translated as a fusion protein Scr-S. However, when cutting at the down stream one, the left 4bp in intron would result in a frameshift, thus only the forward exon can express named Scr-L.
- dCas9 CRISPR interface system:
CRISPR shorts for Clustered Regularly Interspaced Palindromaic Repeats system, which can be targeted to DNA using RNA, enabling genetic editing of any region of the genome in many organisms.[Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease .] In the type II CRISPR/Cas system, a ribonucleoprotein complex formed from a single protein (Cas9), a crRNA, and a trans-acting CRISPR RNA (tracrRNA) can carry out efficient crRNA-directed recognition and site-specific cleavage of foreign DNA(CRISPR RNA maturation by transencoded small RNA and host factor RNase III.). After mutated the endonuclease domains of the Cas9 protein, it creates a programmable RNA-dependent DNA-binding protein. The sgRNA consists of three domains: a 20 nt complementary region for specific DNA binding, a 42 nt hairpin for Cas9 binding (Cas9 handle), and a 40 nt transcription terminator derived from S. Pyogenes. After translation, the Cas9 binds to sgRNA to form a protein-RNA complex, which can recognize target sites in the genome sequence and bind to it. Then, it could block RNA polymerase and transciption elongation.
 "
"