Team:SydneyUni Australia/Project/SinceHK
From 2013.igem.org
(Difference between revisions)
Jbergfield (Talk | contribs) (Created page with "{{Team:SydneyUni_Australia/Style}} {{Team:SydneyUni_Australia/Header}} {{Team:SydneyUni_Australia/Footer}}") |
|||
| Line 3: | Line 3: | ||
| + | __NOTOC__ | ||
| + | =='''Design'''== | ||
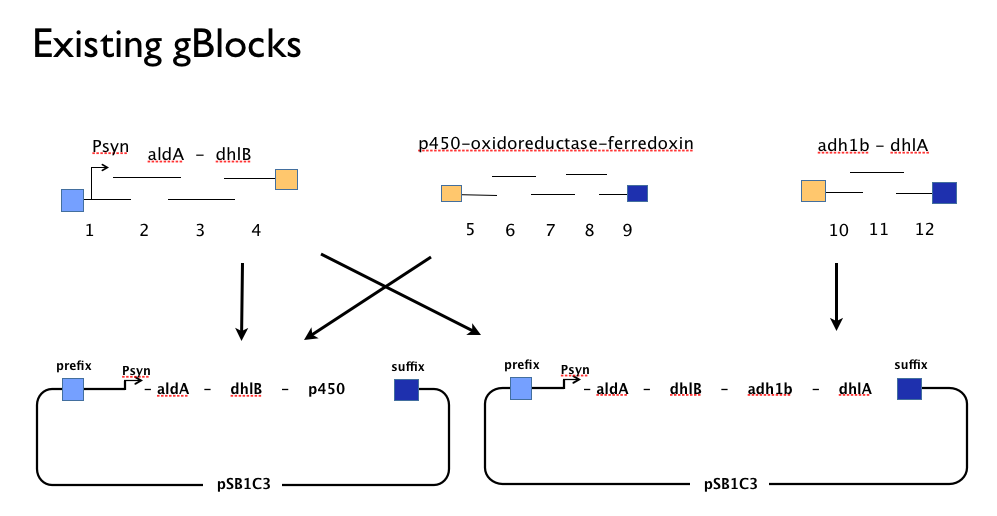
| + | Throughout the year we’d been trying to assemble our DCA-degrading pathway in a single Gibson Assembly reaction. We had trouble. We believe that the strong constitutive expression of some of our parts may harm the cells and effectively create a screen for misassembled Gibson inserts. We also had no flexibility in the face of trouble because we’d designed our gBlocks to assemble all at once, and when this failed we couldn’t assemble the genes in our pathway from parts. | ||
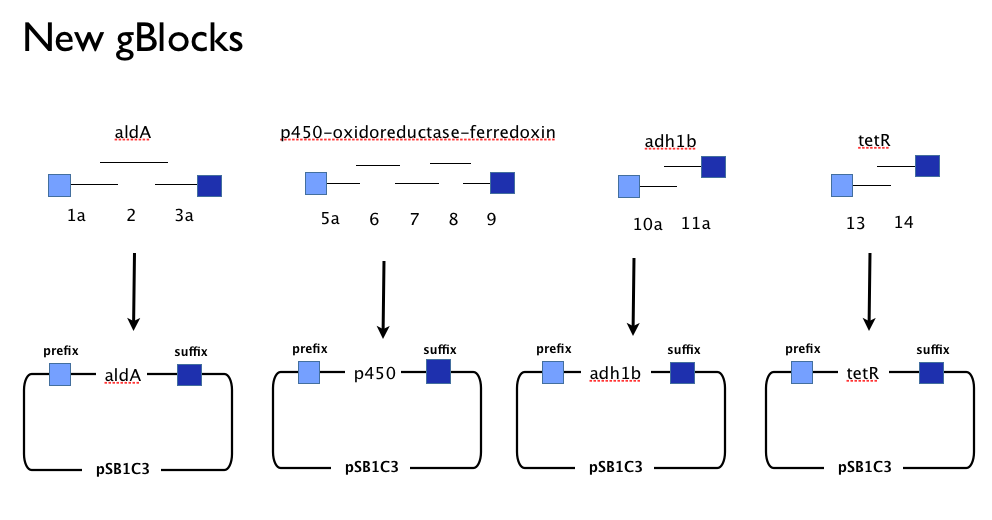
| + | We designed new gBlocks to complement our existing gBlocks, allowing us to assemble each gene in the pathway by itself in the Registry’s shipping vector, pSB1C3. | ||
| + | |||
| + | [[File:2013SydneyUniAustraliaexisting_gBlocks.png|center]] | ||
| + | [[File:2013SydneyUniAustralianew_gBlocks.png|center]] | ||
| + | |||
| + | |||
| + | This approach would allow us to: | ||
| + | *use the more time-consuming but flexible of conventional BioBrick assembly. | ||
| + | *identify which, if any, genes present a toxic or metabolic burden to E. coli. | ||
| + | *characterise each gene individually to inform our model. | ||
| + | *optimise the order of genes in our pathway. | ||
| + | *submit all of our novel genes as characterised parts that can be used by others and in other projects. | ||
| + | |||
| + | |||
| + | =='''Results'''== | ||
| + | |||
| + | For the Friar Tuck, | ||
| + | |||
| + | Talk about the magic we made - | ||
| + | *GA of parts | ||
| + | *cloning with inducible system | ||
| + | *two p450 clones look promising on cl- assay (the GC experiment had way higher conc. tet and way less time with tet) | ||
| + | *...yet to be characterised... | ||
| + | |||
| + | Rob | ||
{{Team:SydneyUni_Australia/Footer}} | {{Team:SydneyUni_Australia/Footer}} | ||
Revision as of 08:36, 27 October 2013


Design
Throughout the year we’d been trying to assemble our DCA-degrading pathway in a single Gibson Assembly reaction. We had trouble. We believe that the strong constitutive expression of some of our parts may harm the cells and effectively create a screen for misassembled Gibson inserts. We also had no flexibility in the face of trouble because we’d designed our gBlocks to assemble all at once, and when this failed we couldn’t assemble the genes in our pathway from parts.
We designed new gBlocks to complement our existing gBlocks, allowing us to assemble each gene in the pathway by itself in the Registry’s shipping vector, pSB1C3.
This approach would allow us to:
- use the more time-consuming but flexible of conventional BioBrick assembly.
- identify which, if any, genes present a toxic or metabolic burden to E. coli.
- characterise each gene individually to inform our model.
- optimise the order of genes in our pathway.
- submit all of our novel genes as characterised parts that can be used by others and in other projects.
Results
For the Friar Tuck,
Talk about the magic we made -
- GA of parts
- cloning with inducible system
- two p450 clones look promising on cl- assay (the GC experiment had way higher conc. tet and way less time with tet)
- ...yet to be characterised...
Rob
 "
"