Team:TU-Eindhoven/Description
From 2013.igem.org
(Created page with "{{:Team:TU-Eindhoven/Template:Base}} {{:Team:TU-Eindhoven/Template:MenuBar}} =Project Description= {{:Team:TU-Eindhoven/Template:FloatingImage | position=left | size=6 | filena...") |
JacquesErnes (Talk | contribs) (→Project Description) |
||
| (16 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
=Project Description= | =Project Description= | ||
| - | {{:Team:TU-Eindhoven/Template:FloatingImage | position= | + | {{:Team:TU-Eindhoven/Template:FloatingImage | position=right | size=6 | filename=genDiagramI.png}} |
| - | Our project focuses on a relatively new form of {{:Team:TU-Eindhoven/Template:Tooltip | text=MRI | tooltip=Magnetic Resonance Imaging }}: {{:Team:TU-Eindhoven/Template:Tooltip | text=CEST | tooltip=Chemical Exchange Saturation Transfer }} imaging. CEST imaging proteins contain hydrogen atoms which can be used to create the same image quality as when conventional heavy metals are used. We use Escherichia | + | Our project focuses on a relatively new form of {{:Team:TU-Eindhoven/Template:Tooltip | text=MRI | tooltip=Magnetic Resonance Imaging }}: {{:Team:TU-Eindhoven/Template:Tooltip | text=CEST | tooltip=Chemical Exchange Saturation Transfer }} imaging. CEST imaging proteins contain hydrogen atoms which can be used to create the same image quality as when conventional heavy metals are used. We use ''Escherichia Coli'' as the [[Team:TU-Eindhoven/Chasis |'''CHASSIS''']] to express CEST proteins that lead to CEST contrast when the bacteria sense a {{:Team:TU-Eindhoven/Template:Tooltip | text=hypoxic | tooltip=Low oxygen saturation }} environment, thus working as a production factory and delivery system for the CEST MRI contrast agent. |
| - | We use ten | + | We use ten [[Team:TU-Eindhoven/Production |'''PROTEINS''']], all of which are rich in Lysine and Arginine amino acids, which leads to a high CEST contrast. In order to achieve the production of the CEST MRI contrast agent by our bacteria, we must create a series of diverse [[Team:TU-Eindhoven/Modeling | '''MODELS''']] focusing on the different components of the MRI contrast agent production. |
| - | In | + | In the [[Team:TU-Eindhoven/Wetlab | '''LAB''']] we produced the CEST MRI proteins aerobically. To enable anaerobic expression, a specialized promoter was designed to react to change in oxygen saturation, thereby triggering the protein expression. After aerobic protein expression the proteins were tested for the quality of their contrast in an MRI machine. |
| - | + | <!--{{:Team:TU-Eindhoven/Template:FloatingImage | position=left | size=2 | filename=bacteriaMiner.png}}<br>--> | |
| + | Based on the principle of our project, we propose two [[Team:TU-Eindhoven/Applications | '''APPLICATIONS''']], tumor CEST MR Imaging and tracking of bacteria in bacterial infections studies. After image acquisition our CEST protein production device can be deactivated and eliminated using the HSV Thymidine kinase/Ganciclovir system as a [[Team:TU-Eindhoven/KillingMechanism | '''KILLING MECHANISM''']]. | ||
| - | + | One of the most prominent threats to our project is the attitude of [[Team:TU-Eindhoven/Outreach|'''SOCIETY''']], which often has trouble coping with the idea of using live bacteria in living patients. Therefore, as our [[Team:TU-Eindhoven/Society|'''HUMAN PRACTICES PROJECT''']], we developed an online source of knowledge related to the explanation of common misconceptions within the field of synthetic biology. However, general safety, not only of our device, but also our own personal safety whilst participating is also important, and thus we take [[Team:TU-Eindhoven/Safety|'''SAFETY''']] issues into consideration in our project. | |
| - | |||
| - | + | ||
{{:Team:TU-Eindhoven/Template:BaseFooter}} | {{:Team:TU-Eindhoven/Template:BaseFooter}} | ||
Latest revision as of 10:05, 26 October 2013



Project Description
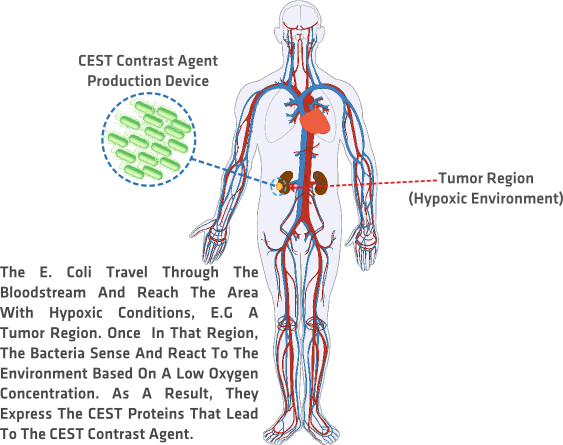
Our project focuses on a relatively new form of MRI: CEST imaging. CEST imaging proteins contain hydrogen atoms which can be used to create the same image quality as when conventional heavy metals are used. We use Escherichia Coli as the CHASSIS to express CEST proteins that lead to CEST contrast when the bacteria sense a hypoxic environment, thus working as a production factory and delivery system for the CEST MRI contrast agent.
We use ten PROTEINS, all of which are rich in Lysine and Arginine amino acids, which leads to a high CEST contrast. In order to achieve the production of the CEST MRI contrast agent by our bacteria, we must create a series of diverse MODELS focusing on the different components of the MRI contrast agent production.
In the LAB we produced the CEST MRI proteins aerobically. To enable anaerobic expression, a specialized promoter was designed to react to change in oxygen saturation, thereby triggering the protein expression. After aerobic protein expression the proteins were tested for the quality of their contrast in an MRI machine.
Based on the principle of our project, we propose two APPLICATIONS, tumor CEST MR Imaging and tracking of bacteria in bacterial infections studies. After image acquisition our CEST protein production device can be deactivated and eliminated using the HSV Thymidine kinase/Ganciclovir system as a KILLING MECHANISM.
One of the most prominent threats to our project is the attitude of SOCIETY, which often has trouble coping with the idea of using live bacteria in living patients. Therefore, as our HUMAN PRACTICES PROJECT, we developed an online source of knowledge related to the explanation of common misconceptions within the field of synthetic biology. However, general safety, not only of our device, but also our own personal safety whilst participating is also important, and thus we take SAFETY issues into consideration in our project.
 "
"



