Team:TU-Eindhoven/Modeling
From 2013.igem.org
| Line 35: | Line 35: | ||
{{:Team:TU-Eindhoven/Template:FloatEnd | caption=General Models of Processes Happening Inside the E. Coli. | id=eColiModelsFigure }} | {{:Team:TU-Eindhoven/Template:FloatEnd | caption=General Models of Processes Happening Inside the E. Coli. | id=eColiModelsFigure }} | ||
| - | As previously mentioned, {{:Team:TU-Eindhoven/Template:Figure | id=eColiModelsFigure }} aims to schematically depict all the models that are related to the diverse components of our device, e.g the proteins that will be produced by the device, the FNR promotor and decoy sites which are related to the gene expression, and the CEST protein production directly related to the CEST MRI contrast agent. To define which proteins are most suitable for the production of the MRI CEST contrast agent by the bacteria we used [[Team:TU-Eindhoven/ProteinSelection | molecular dynamics simulations]]. By using this approach we were able to determine the list of proteins that would be generated by our device, based on the the accessibility of the various exchangable hydrogen atoms of the Arginine and Lysine amino acids of the proteins being considered. In what concerns the FNR promotor and decoy sites, both deterministic models and stochastic simulations were used to predict the concentration of transcription factor and influence of introducing decoy sites. Aside from the individual models made for these two components of the device, a merged deterministic model was created to analyse the complete gene expression process. Sentivity analysis was used on the merged model to determine which elements of the model have the greatest influence on the outcome. Similarly, a PLE analysis was utilized on the decoy sites model. | + | As previously mentioned, {{:Team:TU-Eindhoven/Template:Figure | id=eColiModelsFigure }} aims to schematically depict all the models that are related to the diverse components of our device, e.g the proteins that will be produced by the device, the FNR promotor and decoy sites which are related to the gene expression, and the CEST protein production directly related to the CEST MRI contrast agent. |
| + | |||
| + | To define which proteins are most suitable for the production of the MRI CEST contrast agent by the bacteria we used [[Team:TU-Eindhoven/ProteinSelection | molecular dynamics simulations]]. By using this approach we were able to determine the list of proteins that would be generated by our device, based on the the accessibility of the various exchangable hydrogen atoms of the Arginine and Lysine amino acids of the proteins being considered. In what concerns the FNR promotor and decoy sites, both deterministic models and stochastic simulations were used to predict the concentration of transcription factor and influence of introducing decoy sites. Aside from the individual models made for these two components of the device, a merged deterministic model was created to analyse the complete gene expression process. Sentivity analysis was used on the merged model to determine which elements of the model have the greatest influence on the outcome. Similarly, a PLE analysis was utilized on the decoy sites model. Furthermore, a deterministic model was developed for analyzing the protein production and degradation. Using this information it was possible to predict the average concentration of the CEST proteins produced by our device. These results where then compared to the experimental data. | ||
Revision as of 14:58, 4 September 2013



Modeling Summary
The main objective of the modeling component of our project is to provide insight to the rest of team in what concerns the effect of altering the necessary conditions for the proper operation of each component of our device. Keeping this in mind we established a series of questions that if answered thoroughly would benefit our project. We categorized these questions in four main topics:
Protein Selection
- Which proteins that can be expressed in bacteria are best suited as CEST based markers?
Gene expression
- How does the oxygen concentration affect the concentration of the transcription activator (FNR) in its active form?
- How does introducing decoy sites influence the gene expression rate?
CEST MRI Contrast agent
- What is the average protein concentration produced by the bacteria?
- What the estimated CEST contrast given by the bacteria?
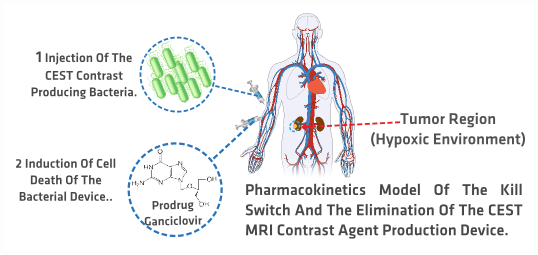
Tumor Imaging In Vivo
- How do bacteria distribute within the human body?
- How long does it take for the bacteria to reach the target area?
- How do the bacteria react to the activation of the killing mechanism?
- What is the bacteria elimination rate?
Based on the previous question list we developed five models that can be divided into two categories, processes occurring inside the bacteria and the interaction of the bacteria with its surroundings. presents a schematic diagram of the developed models related to the first category, while depicts the model that accounts for the interaction between the bacteria and its surroundings when introduced into the human body for tumor imaging. Both figures will be further explained in the next paragraphs. The approaches used for the models range from molecular dynamics simulations to deterministic models and stochastic simulations as well as pharmacokinetics.
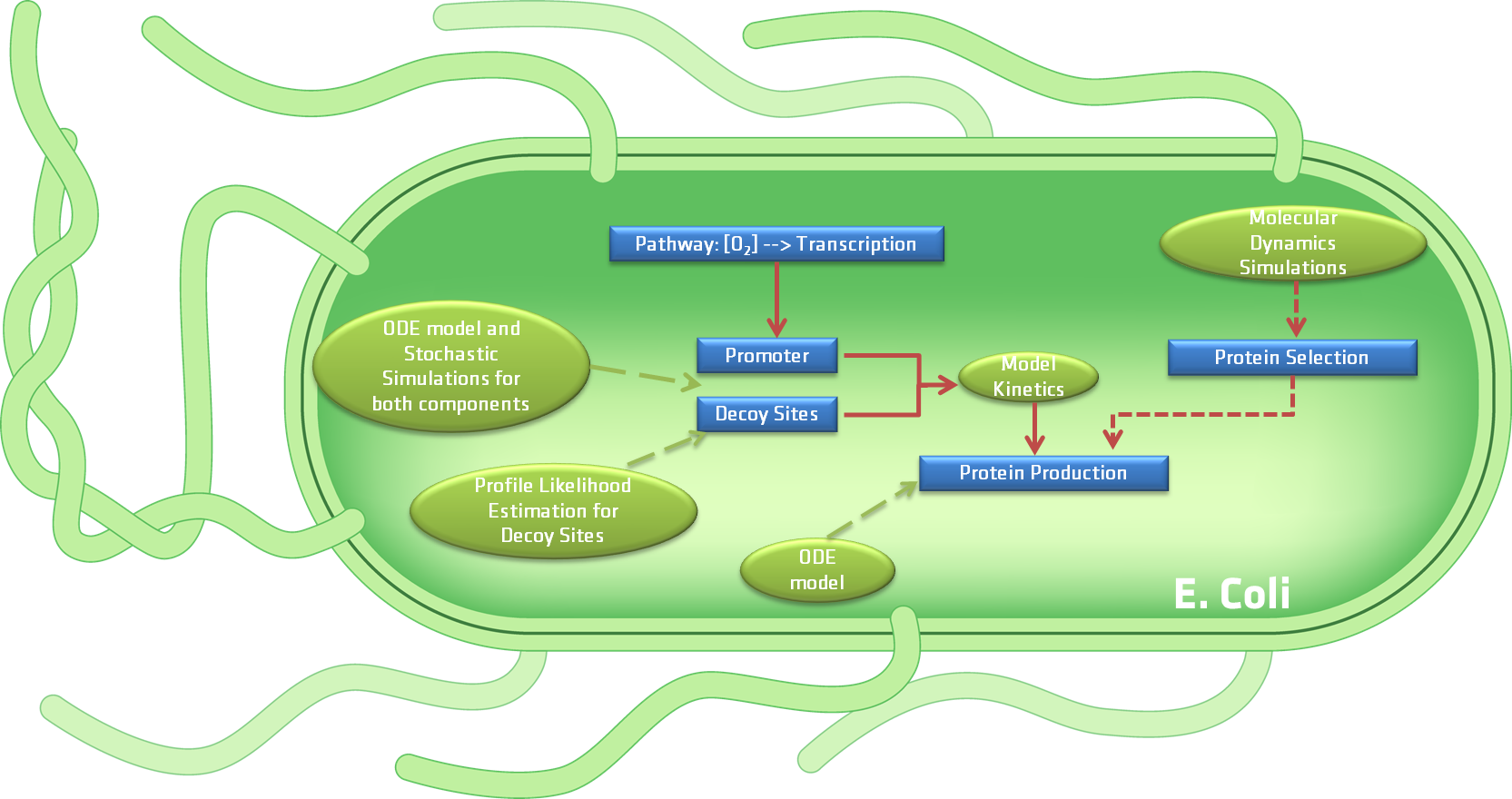
As previously mentioned, aims to schematically depict all the models that are related to the diverse components of our device, e.g the proteins that will be produced by the device, the FNR promotor and decoy sites which are related to the gene expression, and the CEST protein production directly related to the CEST MRI contrast agent.
To define which proteins are most suitable for the production of the MRI CEST contrast agent by the bacteria we used molecular dynamics simulations. By using this approach we were able to determine the list of proteins that would be generated by our device, based on the the accessibility of the various exchangable hydrogen atoms of the Arginine and Lysine amino acids of the proteins being considered. In what concerns the FNR promotor and decoy sites, both deterministic models and stochastic simulations were used to predict the concentration of transcription factor and influence of introducing decoy sites. Aside from the individual models made for these two components of the device, a merged deterministic model was created to analyse the complete gene expression process. Sentivity analysis was used on the merged model to determine which elements of the model have the greatest influence on the outcome. Similarly, a PLE analysis was utilized on the decoy sites model. Furthermore, a deterministic model was developed for analyzing the protein production and degradation. Using this information it was possible to predict the average concentration of the CEST proteins produced by our device. These results where then compared to the experimental data.
 "
"