Team:Warsaw/BiFC Toolbox
From 2013.igem.org
(→Genetic part – The BiFC Toolbox) |
Annamiscicka (Talk | contribs) (→Genetic part – The BiFC Toolbox) |
||
| (16 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
==Genetic part – The BiFC Toolbox== | ==Genetic part – The BiFC Toolbox== | ||
| - | GFP is the most popular fluorescent protein. In 2006 Pédelacq ''et al.'' | + | GFP is the most popular fluorescent protein. In 2006 Pédelacq ''et al.'' engineered and characterized new form of GFP – superfolder GFP (sfGFP), which is more stable and has stronger fluorescence than wild form GFP. We aimed to create other superfolder fluorescent proteins and we succeeded. |
| - | Changing color of fluorescent proteins is possible using | + | Changing color of fluorescent proteins is possible using direct mutagenesis technique. We did it using PCR, with [http://parts.igem.org/Part:BBa_I746908 sfGFP] from Parts Registry (BBa_I746908) as a template. |
| - | * sfBFP (K1093000) - contains Y66H point mutation compared to the original sfGFP | + | * [http://parts.igem.org/Part:BBa_K1093000 sfBFP] (K1093000) - blue fluorescence, contains Y66H point mutation compared to the original sfGFP |
| - | * sfYFP (K1093001) - contains T203Y point mutation compared to the original sfGFP | + | * [http://parts.igem.org/Part:BBa_K1093001 sfYFP] (K1093001) - yellow fluorescence, contains T203Y point mutation compared to the original sfGFP |
| - | * sfCFP (K1093002) - contains Y66W point mutation compared to the original sfGFP | + | * [http://parts.igem.org/Part:BBa_K1093002 sfCFP] (K1093002) - cyan fluorescence, contains Y66W point mutation compared to the original sfGFP |
| - | By doing this we improved sfGFP (by expanding the range of possible excitation/emission optima). One could say that we improved standard forms of CFP, BFP and YFP by creating their superfolder forms. We sent sfBFP, sfYFP and sfCFP to Parts Registry. | + | By doing this we improved sfGFP (by expanding the range of possible excitation/emission optima). One could say that we improved standard forms of CFP, BFP and YFP by creating their superfolder forms. We sent '''sfBFP''', '''sfYFP''' and '''sfCFP''' to Parts Registry. |
| - | When we confirmed the | + | When we confirmed the correctness of our constructs by sequencing, we cloned them on pSB1A3 plasmid, added J23100 promoter and B0034 RBS and subsequently measured them in RF. We chose RF, because our standard medium, LB, displays high internal fluorescence, which makes it impossible to visualize sfBFP in liquid without significant experimental error. |
Excitation/emission maxima for each protein: | Excitation/emission maxima for each protein: | ||
| Line 20: | Line 20: | ||
* sfYFP – 503/540 nm | * sfYFP – 503/540 nm | ||
| - | [[File:Superfolderwykres.jpg|center | + | [[File:Superfolderwykres.jpg|center|border|caption]] |
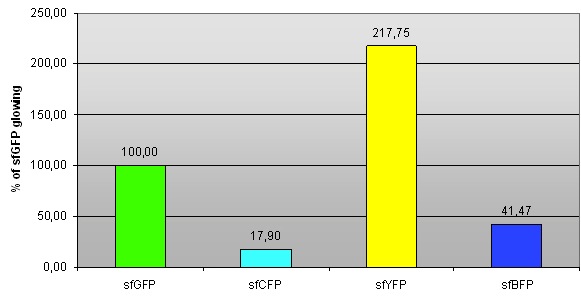
| - | '' | + | ''Fig 1. – Fluorescence of sfCFP, sfYFP and sfBFP compared to sfGFP (BBa_I746908).'' |
| - | We compared fluorescence levels of sfGFP (the fluorescence intensity is normalized, hence | + | We compared fluorescence levels to that of sfGFP (the fluorescence intensity is normalized, hence sfGFP has 100% fluorescence). |
{| class="wikitable" | {| class="wikitable" | ||
| Line 30: | Line 30: | ||
! colspan="3" | Samples | ! colspan="3" | Samples | ||
! scope="col" | Arithmetic mean | ! scope="col" | Arithmetic mean | ||
| - | ! scope="col" | | + | ! scope="col" | Standard deviation |
|- | |- | ||
|sfGFP | |sfGFP | ||
| Line 63: | Line 63: | ||
''Tab 1. – Fluorescence of sfCFP, sfYFP and sfBFP compared to sfGFP (BBa_I746908).'' | ''Tab 1. – Fluorescence of sfCFP, sfYFP and sfBFP compared to sfGFP (BBa_I746908).'' | ||
| - | We found in Parts Registry yellow fluorescent protein, named SYFP2 (super yellow fluorescent protein 2; BBa_ K864100). We decided to compare it with our sfYFP. We | + | We found in Parts Registry a yellow fluorescent protein, named [http://parts.igem.org/Part:BBa_K864100 SYFP2] (super yellow fluorescent protein 2; BBa_ K864100). We decided to compare it with our sfYFP. We cloned BBa_K864100 on pSB1A3 plasmid with J23100 promoter and B0034 RBS, and measured it in RF. Excitation/emission maxima: 503/540 nm. |
| - | [[File:Sfyfpsyfp2wykres.jpg|center | + | [[File:Sfyfpsyfp2wykres.jpg|center|border|caption]] |
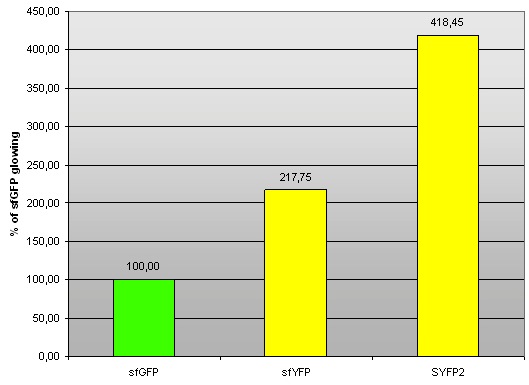
| - | '' | + | ''Fig 2. – Fluorescence of sfYFP and SYFP2 compared to sfGFP (BBa_I746908).'' |
{| class="wikitable" | {| class="wikitable" | ||
| Line 73: | Line 73: | ||
! colspan="3" | Samples | ! colspan="3" | Samples | ||
! scope="col" | Arithmetic mean | ! scope="col" | Arithmetic mean | ||
| - | ! scope="col" | | + | ! scope="col" | Standard deviation |
|- | |- | ||
|sfGFP | |sfGFP | ||
| Line 99: | Line 99: | ||
''Tab 2. – Fluorescence of sfYFP and SYFP2 compared to sfGFP (BBa_I746908).'' | ''Tab 2. – Fluorescence of sfYFP and SYFP2 compared to sfGFP (BBa_I746908).'' | ||
| - | The fact that fluorescence of sfYFP is not as strong as fluorescence of SYFP2 is | + | The fact that fluorescence of sfYFP is not as strong as fluorescence of SYFP2 is slighlty disappointing. However, we sincerely congratulate iGEM2012 Uppsala Team for creating a great yellow fluorescent protein. |
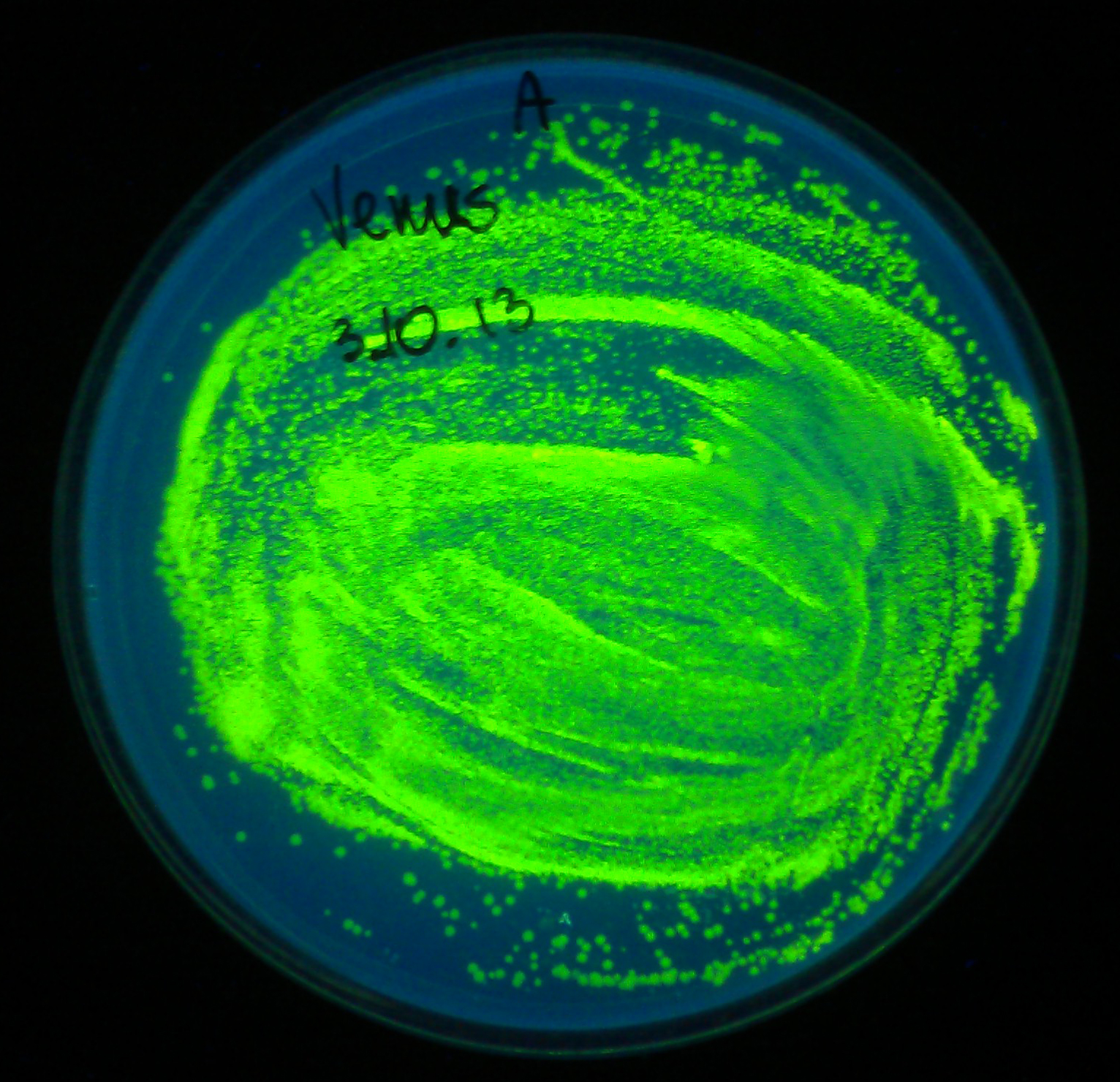
| - | Comparison with sfVenus (and sfCerulean) protein which we also designed this year would be most interesting. Unfortunately, the synthetic construct did not reach us in time before wiki freeze. | + | Comparison with sfVenus (and sfCerulean) protein which we also designed this year would be most interesting. Unfortunately, the synthetic construct did not reach us in time before wiki freeze. W did however manage to confirm the correct functioning of regular Venus protein (although not quantitatively), codon optimised for ''E. coli'' (K1093032) |
| - | We created superfolder BFP, CFP and YFP with the intention of creating a “BiFC Toolbox” for synthetic biology. BiFC (Bimolecular Fluorescent Complementation) is method used to validate and visualize protein-protein interactions in living cells. Fragments of fluorescent proteins are fused to proteins that we study and if they interact, functional fluorescent protein is formed and a | + | |
| + | [[File:Venus_optimised.jpg|400px|thumb|center|alt=A|Codon-optimised Venus]] | ||
| + | |||
| + | |||
| + | We created superfolder BFP, CFP and YFP with the intention of creating a '''“BiFC Toolbox”''' for synthetic biology. BiFC (Bimolecular Fluorescent Complementation) is a method used to validate and visualize protein-protein interactions in living cells. Fragments of fluorescent proteins are fused to proteins that we study and if they interact, functional fluorescent protein is formed and a fluorescent signal is emitted. | ||
Superfolder proteins are perfect for BiFC system due to their improved stability and relatively fast folding kinetics. | Superfolder proteins are perfect for BiFC system due to their improved stability and relatively fast folding kinetics. | ||
| - | We truncated sfGFP, sfCFP, sfBFP and sfYFP by PCR using specific primers. N-terminal fragment is long (645 bp) and specific to each protein. C-terminal fragment is very short (54 bp) and comes from sfGFP, but we suspect that it would also work well with other fluorescent proteins as well. C- | + | We truncated sfGFP, sfCFP, sfBFP and sfYFP by PCR using specific primers. N-terminal fragment is long (645 bp) and specific to each protein. C-terminal fragment is very short (54 bp) and comes from sfGFP, but we suspect that it would also work well with other fluorescent proteins as well. C-terminus is designed in two variants: m6 and m12 (differing in sensitivity to specificity ratio). |
| - | Moreover, we attempted to produce BiFC fragments from GFP (E0040), mCherry (J06504) and mOrange (E2050). | + | Moreover, we attempted to produce BiFC fragments from [http://parts.igem.org/Part:BBa_E0040 GFP] (E0040), [http://parts.igem.org/Part:BBa_J06504 mCherry] (J06504) and [http://parts.igem.org/Part:BBa_E2050 mOrange] (E2050). |
| - | We | + | We successfully amplificated '''N-sfBFP, N-sfYFP, N-sfCFP, N-mCherry, C-mCherry''' and cloned them into pSB1C3 plasmid. We sent those parts to Parts Registry. C-m6 and C-m12 fragments, in part due to their small size, proved to be difficult to clone. Thus, we didn’t manage to submit the DNA samples before the deadline. |
| - | To verify the functionality of our BiFC fragments, we decided to use b-Fos and b-Jun proteins, | + | To verify the functionality of our BiFC fragments, we decided to use '''b-Fos''' and '''b-Jun''' proteins, which interact with each other. We planned to fuse b-Jun with N-terminal fragment and b-Fos with C-terminal one. Furthermore, we intended to use b-Fos without leucine zipper (in this form it is not able to interact with b-Jun) as a negative control. For measurements we lysed bacteria expressing either b-Fos and b-Jun fused with part of a fluorescent protein and mixed the lysates. This way we will create not only a wide range of BiFC proteins of different parameters, but also provide systematic and standardized information about sensitivity and specificity of each combination. We will also measure Venus- and Cerulean-based BiFC fragments already present in the registry. |
Unfortunately, vacation is over and we did not manage to fulfill our ultimate goal and measure our constructs this year. We will however continue working on the toolbox, as we feel it will prove a valuable addition to the Registry. | Unfortunately, vacation is over and we did not manage to fulfill our ultimate goal and measure our constructs this year. We will however continue working on the toolbox, as we feel it will prove a valuable addition to the Registry. | ||
| Line 116: | Line 120: | ||
'''References:''' | '''References:''' | ||
| - | + | # Pédelacq, J.-D., Cabantous, S., Tran, T., Terwilliger, T. C. & Waldo, G. S. Engineering and characterization of a superfolder green fluorescent protein. Nature biotechnology 24, 79–88 (2006). | |
| - | + | # Shyu, Y. J., Liu, H., Deng, X., Hu, C. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. BioTechniques 40:61-66 (2006). | |
| - | + | ||
| - | + | ||
{{:Team:Warsaw/Templates/StandardPageEnd}} | {{:Team:Warsaw/Templates/StandardPageEnd}} | ||
Latest revision as of 03:14, 5 October 2013
BiFC Toolbox
Genetic part – The BiFC Toolbox
GFP is the most popular fluorescent protein. In 2006 Pédelacq et al. engineered and characterized new form of GFP – superfolder GFP (sfGFP), which is more stable and has stronger fluorescence than wild form GFP. We aimed to create other superfolder fluorescent proteins and we succeeded. Changing color of fluorescent proteins is possible using direct mutagenesis technique. We did it using PCR, with sfGFP from Parts Registry (BBa_I746908) as a template.
- sfBFP (K1093000) - blue fluorescence, contains Y66H point mutation compared to the original sfGFP
- sfYFP (K1093001) - yellow fluorescence, contains T203Y point mutation compared to the original sfGFP
- sfCFP (K1093002) - cyan fluorescence, contains Y66W point mutation compared to the original sfGFP
By doing this we improved sfGFP (by expanding the range of possible excitation/emission optima). One could say that we improved standard forms of CFP, BFP and YFP by creating their superfolder forms. We sent sfBFP, sfYFP and sfCFP to Parts Registry.
When we confirmed the correctness of our constructs by sequencing, we cloned them on pSB1A3 plasmid, added J23100 promoter and B0034 RBS and subsequently measured them in RF. We chose RF, because our standard medium, LB, displays high internal fluorescence, which makes it impossible to visualize sfBFP in liquid without significant experimental error.
Excitation/emission maxima for each protein:
- sfGFP – 485/510 nm
- sfBFP – 380/445 nm
- sfCFP – 425/475 nm
- sfYFP – 503/540 nm
Fig 1. – Fluorescence of sfCFP, sfYFP and sfBFP compared to sfGFP (BBa_I746908).
We compared fluorescence levels to that of sfGFP (the fluorescence intensity is normalized, hence sfGFP has 100% fluorescence).
| - | Samples | Arithmetic mean | Standard deviation | ||
|---|---|---|---|---|---|
| sfGFP | 100,60 | 100,37 | 99,02 | 100,00 | 0,85 |
| sfCFP | 17,95 | 18,38 | 17,36 | 17,90 | 0,51 |
| sfBFP | 40,65 | 41,52 | 42,25 | 41,47 | 0,80 |
| sfYFP | 218,87 | 216,51 | 217,87 | 217,75 | 1,19 |
Tab 1. – Fluorescence of sfCFP, sfYFP and sfBFP compared to sfGFP (BBa_I746908).
We found in Parts Registry a yellow fluorescent protein, named SYFP2 (super yellow fluorescent protein 2; BBa_ K864100). We decided to compare it with our sfYFP. We cloned BBa_K864100 on pSB1A3 plasmid with J23100 promoter and B0034 RBS, and measured it in RF. Excitation/emission maxima: 503/540 nm.
Fig 2. – Fluorescence of sfYFP and SYFP2 compared to sfGFP (BBa_I746908).
| - | Samples | Arithmetic mean | Standard deviation | ||
|---|---|---|---|---|---|
| sfGFP | 100,60 | 100,37 | 99,02 | 100,00 | 0,85 |
| sfYFP | 218,87 | 216,51 | 217,87 | 217,75 | 1,19 |
| SYFP2 | 424,70 | 418,26 | 412,40 | 418,45 | 6,15 |
Tab 2. – Fluorescence of sfYFP and SYFP2 compared to sfGFP (BBa_I746908).
The fact that fluorescence of sfYFP is not as strong as fluorescence of SYFP2 is slighlty disappointing. However, we sincerely congratulate iGEM2012 Uppsala Team for creating a great yellow fluorescent protein. Comparison with sfVenus (and sfCerulean) protein which we also designed this year would be most interesting. Unfortunately, the synthetic construct did not reach us in time before wiki freeze. W did however manage to confirm the correct functioning of regular Venus protein (although not quantitatively), codon optimised for E. coli (K1093032)
We created superfolder BFP, CFP and YFP with the intention of creating a “BiFC Toolbox” for synthetic biology. BiFC (Bimolecular Fluorescent Complementation) is a method used to validate and visualize protein-protein interactions in living cells. Fragments of fluorescent proteins are fused to proteins that we study and if they interact, functional fluorescent protein is formed and a fluorescent signal is emitted.
Superfolder proteins are perfect for BiFC system due to their improved stability and relatively fast folding kinetics.
We truncated sfGFP, sfCFP, sfBFP and sfYFP by PCR using specific primers. N-terminal fragment is long (645 bp) and specific to each protein. C-terminal fragment is very short (54 bp) and comes from sfGFP, but we suspect that it would also work well with other fluorescent proteins as well. C-terminus is designed in two variants: m6 and m12 (differing in sensitivity to specificity ratio). Moreover, we attempted to produce BiFC fragments from GFP (E0040), mCherry (J06504) and mOrange (E2050).
We successfully amplificated N-sfBFP, N-sfYFP, N-sfCFP, N-mCherry, C-mCherry and cloned them into pSB1C3 plasmid. We sent those parts to Parts Registry. C-m6 and C-m12 fragments, in part due to their small size, proved to be difficult to clone. Thus, we didn’t manage to submit the DNA samples before the deadline.
To verify the functionality of our BiFC fragments, we decided to use b-Fos and b-Jun proteins, which interact with each other. We planned to fuse b-Jun with N-terminal fragment and b-Fos with C-terminal one. Furthermore, we intended to use b-Fos without leucine zipper (in this form it is not able to interact with b-Jun) as a negative control. For measurements we lysed bacteria expressing either b-Fos and b-Jun fused with part of a fluorescent protein and mixed the lysates. This way we will create not only a wide range of BiFC proteins of different parameters, but also provide systematic and standardized information about sensitivity and specificity of each combination. We will also measure Venus- and Cerulean-based BiFC fragments already present in the registry.
Unfortunately, vacation is over and we did not manage to fulfill our ultimate goal and measure our constructs this year. We will however continue working on the toolbox, as we feel it will prove a valuable addition to the Registry.
References:
- Pédelacq, J.-D., Cabantous, S., Tran, T., Terwilliger, T. C. & Waldo, G. S. Engineering and characterization of a superfolder green fluorescent protein. Nature biotechnology 24, 79–88 (2006).
- Shyu, Y. J., Liu, H., Deng, X., Hu, C. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. BioTechniques 40:61-66 (2006).
 "
"