Team:Paris Saclay/Notebook/July/4
From 2013.igem.org
(Difference between revisions)
(→Lab work) |
(→Lab work) |
||
| Line 19: | Line 19: | ||
**''BioBrick RBS+amilCP+terminator in plasmid PSB1C3 '' | **''BioBrick RBS+amilCP+terminator in plasmid PSB1C3 '' | ||
| - | <u>Electrophoresis band size estimation</u> | + | <u>Electrophoresis band size estimation</u><br> |
<p>We used Clonemanager for band size estimation:</p> | <p>We used Clonemanager for band size estimation:</p> | ||
<br> | <br> | ||
| Line 59: | Line 59: | ||
<u>Stock BBa_K1155000</u><br> | <u>Stock BBa_K1155000</u><br> | ||
<p>1 ml of the confirmed sample mixed with 500µl glycerol. And we stocked them at -20°C.</p><br> | <p>1 ml of the confirmed sample mixed with 500µl glycerol. And we stocked them at -20°C.</p><br> | ||
| + | |||
<u>Plasmid DNA extraction</u><br> | <u>Plasmid DNA extraction</u><br> | ||
| + | |||
From the cell-culture medium(2 samples plasmid with fnr), we performed some plasmid DNA extraction.<br> | From the cell-culture medium(2 samples plasmid with fnr), we performed some plasmid DNA extraction.<br> | ||
| + | |||
<u>Restriction digest</u><br> | <u>Restriction digest</u><br> | ||
<p>We used 3 enzymes of restriction, they were Not I, Mlu I and Hpa I. And we prepared 2 * 3 = 6 tubes. | <p>We used 3 enzymes of restriction, they were Not I, Mlu I and Hpa I. And we prepared 2 * 3 = 6 tubes. | ||
| - | So we add into each tube:</p> | + | So we add into each tube:</p><br> |
*Extracted DNA solution : 2µl | *Extracted DNA solution : 2µl | ||
*Buffer Oranger : 2µl | *Buffer Oranger : 2µl | ||
| Line 69: | Line 72: | ||
*H2O : 15.5µl | *H2O : 15.5µl | ||
*Total : 20µl | *Total : 20µl | ||
| - | <p>The incubation was at 37°C during 90min</p> | + | <br><p>The incubation was at 37°C during 90min</p> |
Revision as of 23:16, 18 September 2013
Notebook : July 4
Summary:
For regulator system:
- estimated the size of segments for bands of electrophoresis by using Clone Manager.
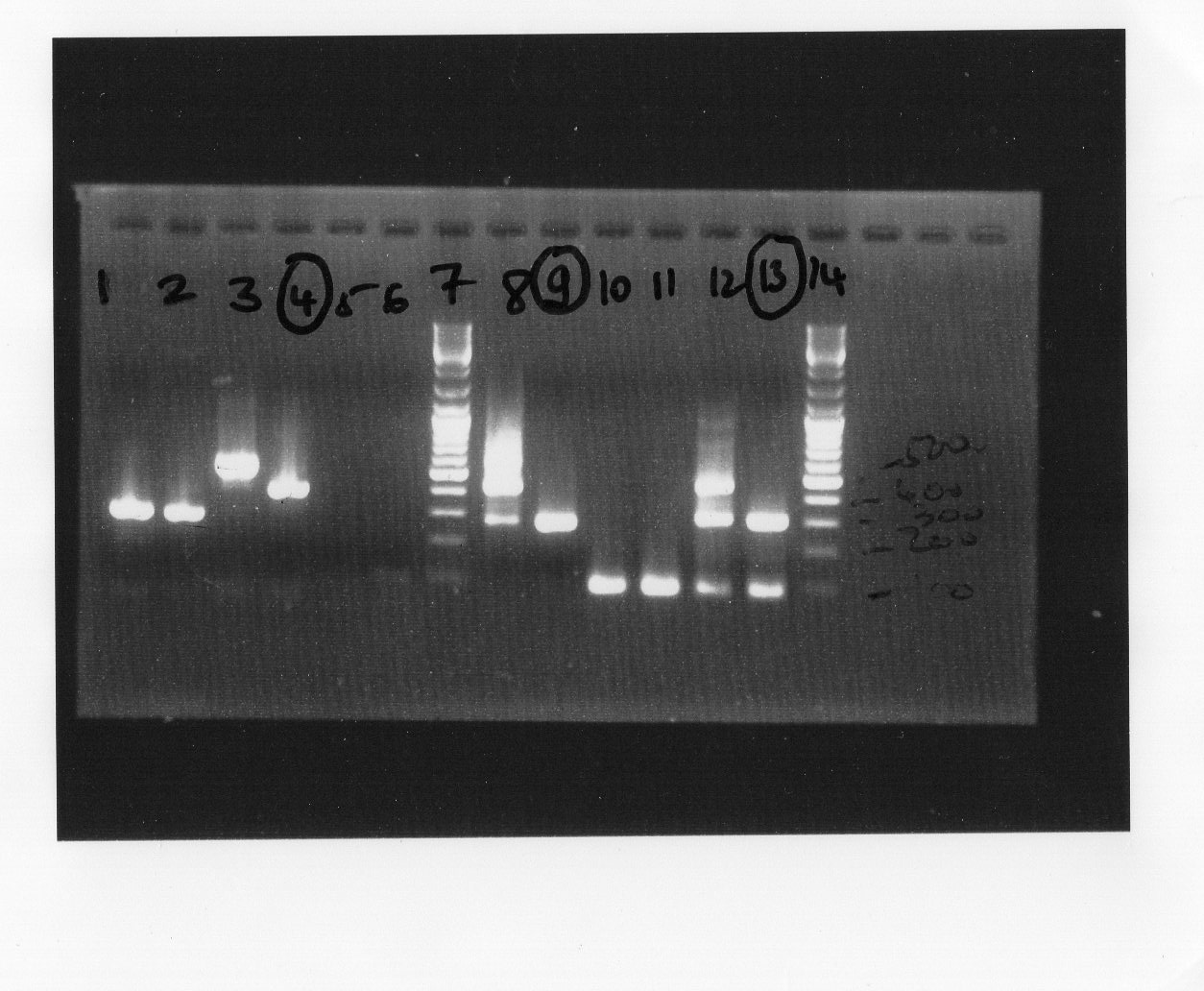
- Made interpretation for electrophoresis. The picture shows that the experimental result was coherent with our estimation. We constructed our first BrioBrick, BBa_K1155000 (fnr+plasmid PSB1C3).
- stored 2 colonies who contain BioBrick BBa_K1155000
- the plasmid DNA extraction was performed for BBa_K1155000.
- The extract of plasmid DNA which contain BBa_K1155000 was digested by Not I, Mlu I, Hpa I, and were amplified by PCR.
- Seeded the 4 additional broths (2 for amilCP, 2 for FNR) for plasmid extraction(mini prep).
For PCBs sensor system:
- Received the bacterial strain: pseudomonas KE707.
Lab work
- A.aero/anaerobic regulation system
- 2.BioBrick RBS+LacZ+terminator in plasmid PSB1C3
- BioBrick RBS+amilCP+terminator in plasmid PSB1C3
Electrophoresis band size estimation
We used Clonemanager for band size estimation:
| molecule | Primer pair | Size |
| Plasmid without fnr | VF/VR | 277bp |
| Plasmid with fnr | Pfnr_up/Pfnr_down | 276bp |
| Plasmid with fnr | Pfnr_up/VR | 311bp |
PCR interpretation
|
We observed for well 10 and well 11 there were a problem which could be some fault in the manipulation. However, the well 4,9,13 conformed to our estimation.
Stock BBa_K1155000
1 ml of the confirmed sample mixed with 500µl glycerol. And we stocked them at -20°C.
Plasmid DNA extraction
From the cell-culture medium(2 samples plasmid with fnr), we performed some plasmid DNA extraction.
Restriction digest
We used 3 enzymes of restriction, they were Not I, Mlu I and Hpa I. And we prepared 2 * 3 = 6 tubes. So we add into each tube:
- Extracted DNA solution : 2µl
- Buffer Oranger : 2µl
- Enzyme : 0.5µl
- H2O : 15.5µl
- Total : 20µl
The incubation was at 37°C during 90min
| Previous day | Back to calendar | Next day |
 "
"