Template:Kyoto/Notebook/Sep 8
From 2013.igem.org
(Difference between revisions)
(→Sep 8) |
(→Gel Extraction) |
||
| Line 16: | Line 16: | ||
|6||RBS-lysis3-DT||XbaI & PstI | |6||RBS-lysis3-DT||XbaI & PstI | ||
|} | |} | ||
| - | [[File: | + | [[File:igku_981b.png]]<br> |
| - | [[File: | + | [[File:igku_981a.png]]<br> |
{| class="wikitable" | {| class="wikitable" | ||
!Name||concentration[µg/mL]||260/280||260/230 | !Name||concentration[µg/mL]||260/280||260/230 | ||
| Line 42: | Line 42: | ||
|10||RBS-lacZα-DT||XbaI & PstI | |10||RBS-lacZα-DT||XbaI & PstI | ||
|} | |} | ||
| - | [[File: | + | [[File:igku_982b.png]]<br> |
| - | [[File: | + | [[File:igku_982a.png]]<br> |
{| class="wikitable" | {| class="wikitable" | ||
!Name||concentration[µg/mL]||260/280||260/230 | !Name||concentration[µg/mL]||260/280||260/230 | ||
| Line 66: | Line 66: | ||
|6||pSB4K5||EcoRI & SpeI | |6||pSB4K5||EcoRI & SpeI | ||
|} | |} | ||
| - | [[File: | + | [[File:igku_983b.png]]<br> |
| - | [[File: | + | [[File:igku_983a.png]]<br> |
{| class="wikitable" | {| class="wikitable" | ||
!Name||concentration[µg/mL]||260/280||260/230 | !Name||concentration[µg/mL]||260/280||260/230 | ||
| Line 96: | Line 96: | ||
|12||pSB1C3||XbaI & PstI | |12||pSB1C3||XbaI & PstI | ||
|} | |} | ||
| - | [[File: | + | [[File:igku_984b.png]]<br> |
| - | [[File: | + | [[File:igku_984a.png]]<br> |
{| class="wikitable" | {| class="wikitable" | ||
!Name||concentration[µg/mL]||260/280||260/230 | !Name||concentration[µg/mL]||260/280||260/230 | ||
Revision as of 04:53, 25 September 2013
Contents |
Sep 8
Gel Extraction
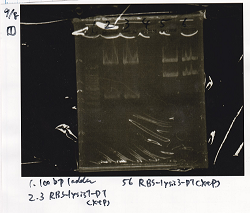
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 100bp ladder | |
| 2 | RBS-lysis1-DT | XbaI & PstI |
| 3 | RBS-lysis1-DT | XbaI & PstI |
| 5 | RBS-lysis3-DT | XbaI & PstI |
| 6 | RBS-lysis3-DT | XbaI & PstI |
| Name | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| RBS-lysis1-DT (XbaI & PstI) | 5.3 | 1.98 | 0.06 |
| RBS-lysis3-DT (XbaI & PstI) | 5.3 | 1.98 | 0.06 |
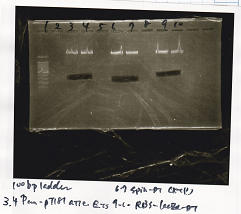
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 100bp ladder | |
| 3 | Pcon-pT181 attenuator | EcoRI & SpeI |
| 4 | Pcon-pT181 attenuator | EcoRI & SpeI |
| 6 | Spinach-DT | XbaI & PstI |
| 7 | Spinach-DT | XbaI & PstI |
| 9 | RBS-lacZα-DT | XbaI & PstI |
| 10 | RBS-lacZα-DT | XbaI & PstI |
| Name | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| Pcon-pT181 attenuator (EcoRI & SpeI) | 5.2 | 1.82 | 0.36 |
| Spinach-DT (XbaI & PstI) | 8.0 | 1.87 | 0.25 |
| RBS-lacZα-DT (XbaI & PstI) | 10.3 | 1.71 | 0.54 |
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 1kbp ladder | |
| 2 | Ptet-pT181 antisense | EcoRI & PstI |
| 3 | Ptet-pT181 antisense | EcoRI & PstI |
| 5 | pSB4K5 | EcoRI & SpeI |
| 6 | pSB4K5 | EcoRI & SpeI |
| Name | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| Ptet-pT181 antisense(EcoRI & PstI) | 30.6 | 1.84 | 1.16 |
| pSB4K5 (EcoRI & SpeI) | 21.8 | 1.87 | 0.98 |
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 1kbp ladder | |
| 2 | Pcon-pT181 attenuator | SpeI & PstI |
| 3 | Pcon-pT181 attenuator | SpeI & PstI |
| 5 | Plac | SpeI & PstI |
| 6 | Plac | SpeI & PstI |
| 8 | pSB1C3 | EcoRI & SpeI |
| 9 | pSB1C3 | EcoRI & SpeI |
| 11 | pSB1C3 | XbaI & PstI |
| 12 | pSB1C3 | XbaI & PstI |
| Name | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| Pcon-pT181 attenuator (SpeI & PstI) | 14.5 | 1.91 | 1.09 |
| Plac (SpeI & PstI) | 11.6 | 1.74 | 1.19 |
| pSB1C3 (EcoRI & SpeI) | 5.7 | 1.74 | 0.34 |
| pSB1C3 (XbaI & PstI) | 5.6 | 2.00 | 0.37 |
Miniprep
| DNA | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| Pcon-pT181 anti | 243.0 | 1.90 | 1.81 |
| Pcon-Spinach-DT | 253.7 | 1.57 | 1.51 |
Ligation
| state | Vector(µL) | Inserter(µL) | Ligation High ver.2 | ||
|---|---|---|---|---|---|
| experiment | 9/8 Plac S+P | 4.3 | 9/3 pT181 attenuator | 3.1 | |
| experiment | 9/8 Plac S+P | 4.3 | 9/7 aptamer12-1R-DT X+P | 2.1 | |
| experiment | 9/8 Plac S+P | 4.3 | 9/8 spinach-DT X+P | 4.6 | |
| experiment | 9/6 Plac S+P | 2 | 9/7 RBS-lysis2-DT X+P | 10 | |
| experiment | 9/8 Plac S+P | 4.3 | 9/7 RBS-lysis2-DT X+P | 10 | |
| experiment | 9/8 Ptet-pT181 anti | 1.6 | 9/8 spinach-DT X+P | 4.3 | |
| experiment | 9/6 Pcon S+P | 1.4 | 9/8 RBS-laczα-DT X+P | 4.8 | |
| experiment | 9/8 Pcon-pT181 attenuator S+P | 3.4 | 9/7 aptamer12-1R-DT X+P | 1.9 | |
| experiment | 9/6 Pcon S+P | 1.4 | 9/7 aptamer12-1R-DT X+P | 2.1 | |
| experiment | 9/8 DT | 3.0 | 9/8 Pcon-pT181 attenuator E+S | 7.9 | |
| experiment | 9/6 Plux S+P | 2 | 9/7 RBS-lysis1-DT X+P | 7.2 | |
| experiment | 9/8 Plac S+P | 4.3 | 9/7 RBS-lysis1-DT X+P | 7.2 | |
| experiment | 9/4 Plac S+P | 1.7 | 9/8 RBS-laczα-DT X+P | 4.8 | |
Restriction Enzyme Digestion
| 9/8 Pcon-pT181 anti | E | S | X | P | BSA | Buffer | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| 2 cuts | 8.2µL | 0µL | 1µL | 0µL | 1µL | 3µL | 3µL | 13.8µL | 30µL |
| NC | 0.4µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL |
| 9/8 Pcon-spin-DT | E | S | X | P | BSA | Buffer | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| 2 cuts | 7.9µL | 1µL | 1µL | 0µL | 0µL | 3µL | 3µL | 14.1µL | 30µL |
| NC | 0.4µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL |
| 8/21 F1 atte | E | S | X | P | BSA | Buffer | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| 2 cuts | 6.5µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 15.5µL | 30µL |
| NC | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL |
| 8/21 F3m2 atte | E | S | X | P | BSA | Buffer | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| 2 cuts | 6.0µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 16µL | 30µL |
| NC | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL |
| 8/21 F1 anti | E | S | X | P | BSA | Buffer | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| 2 cuts | 6.7µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 15.3µL | 30µL |
| NC | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL |
| 8/21 F6 anti | E | S | X | P | BSA | Buffer | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| 2 cuts | 6.0µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 16µL | 30µL |
| NC | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL |
| 8/21 tetR aptamer 12_1M | E | S | X | P | BSA | Buffer | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| 2 cuts | 5.3µL | 1µL | 1µL | 0µL | 0µL | 3µL | 3µL | 16.7µL | 30µL |
| NC | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL |
| 8/21 tetR aptamer 12_P | E | S | X | P | BSA | Buffer | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| 2 cuts | 13.3µL | 1µL | 1µL | 0µL | 0µL | 3µL | 3µL | 8.7µL | 30µL |
| NC | 0.7µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.3µL | 10µL |
| 9/4 spin-DT | E | S | X | P | BSA | Buffer | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| 2 cuts | 5.4µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 16.6µL | 30µL |
| NC | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL |
| 9/7 RBS-lysis1-DT | E | S | X | P | BSA | Buffer | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| 2 cuts | 10µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 12µL | 30µL |
| NC | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL |
| 9/6 RBS-lysis2-DT | E | S | X | P | BSA | Buffer | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| 2 cuts | 13µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 9µL | 30µL |
| NC | 0.7µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.3µL | 10µL |
| 9/7 RBS-lysis3-DT | E | S | X | P | BSA | Buffer | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| 2 cuts | 10µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 12µL | 30µL |
| NC | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL |
| 8/20 J23100 RBS-lacZα-DT-(1) | E | S | X | P | BSA | Buffer | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| 2 cuts | 7µL | 0.5µL | 0.5µL | 0µL | 0µL | 3µL | 3µL | 16µL | 30µL |
| NC | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL |
| 8/20 RBS-lacZα-DT | E | S | X | P | BSA | Buffer | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|
| 2 cuts | 10µL | 0.5µL | 0.5µL | 0µL | 0µL | 3µL | 3µL | 13µL | 30µL |
| NC | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL |
Transformation
| Name | Sample(µL) | Competent Cells(µL) | Total(µL) | Plate |
|---|---|---|---|---|
| 9/8 Pcont+aptamer 12-1R-DT | 1 | 10 | 11 | Amp |
| 9/8 Pcon+RBS-lacZα-DT | 1 | 10 | 11 | Amp |
| 9/8 Pcon-pT181 attenuater+aptamer 12-1R-DT | 1 | 10 | 11 | Amp |
| 9/8 Plac+RBS-lysis1-DT | 2 | 20 | 22 | CP |
| 9/8 Plac+RBS-lysis2-DT | 2 | 20 | 22 | CP |
| 9/8 Plac+aptamer 12-1R-DT | 2 | 20 | 22 | CP |
| 9/8 Plac+spinach | 2 | 20 | 22 | CP |
| 9/8 Plac+pT181 attenuater | 2 | 20 | 22 | CP |
| 9/8 Plux+RBS-lysis1-DT | 1 | 10 | 11 | CP |
| 9/8 Plux+RBS-lysis2-DT | 1 | 10 | 11 | CP |
| 9/8 Ptet+RBS-lacZα-DT | 1 | 10 | 11 | CP |
| 9/8 Ptet-pT181 anti+spinach-DT | 1 | 10 | 11 | CP |
| 9/8 Pcon-pT181 attenuater+DT | 1 | 10 | 11 | CP |
Electrophoresis
| Lane | Sample | Enzyme1 | Enzyme2 |
|---|---|---|---|
| 1 | 100bp ladder | ||
| 2 | Pcon-spin-DT | E | S |
| 3 | Pcon-spin-DT | ||
| 4 | Fusion1 attenuator | X | P |
| 5 | Fusion1 attenuator | ||
| 6 | Fusion3m2 attenuator | X | P |
| 7 | Fusion3m2 attenuator | ||
| 8 | Fusion1 anti | X | P |
| 9 | Fusion1 anti | ||
| 10 | Fusion6 anti | X | P |
| 11 | Fusion6 anti | ||
| 12 | aptamer 12-1M | X | P |
| 13 | aptamer 12-1M | ||
| 14 | aptamer 12-P | X | P |
| 15 | aptamer 12-P | ||
| 16 | spinach-DT | X | P |
| Lane | Sample | Enzyme1 | Enzyme2 |
|---|---|---|---|
| 1 | spinach-DT | ||
| 2 | J23100-RBS-lacZα-DT | E | S |
| 3 | J23100-RBS-lacZα-DT | ||
| 4 | 100bp ladder | ||
| 5 | RBS-lacZα-DT | E | S |
| 6 | RBS-lacZα-DT | ||
| 7 | RBS-lysis1-DT | X | P |
| 8 | RBS-lysis1-DT |
| Lane | Sample | Enzyme1 | Enzyme2 |
|---|---|---|---|
| 1 | 100bp ladder | ||
| 2 | RBS-lysis2-DT | X | P |
| 3 | RBS-lysis2-DT | ||
| 4 | RBS-lysis3-DT | X | P |
| 5 | RBS-lysis3-DT | ||
| 6 | Pcon pT181 anti | S | P |
| 7 | Pcon pT181 anti |
Gel Extraction
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 100bp ladder | |
| 2 | Pcon-spin-DT | E+S |
| 3 | Pcon-spin-DT | E+S |
| 5 | Fusion 1 attenuator | X+P |
| 6 | Fusion 1 attenuator | X+P |
| 8 | Fusion3m2 attenuator | X+P |
| 9 | Fusion3m2 attenuator | X+P |
| 11 | Fusion 1 anti | X+P |
| 12 | Fusion 1 anti | X+P |
File:Igku xxbeforexx.xxx
File:Igku xxafterxx.xxx
| Name | quantity | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|---|
| Pcon-spin-DT(E+S) | 10.5 | 1.86 | 0.39 | |
| Fusion 1 attenuator(X+P) | 7.0 | 1.71 | 0.27 | |
| Fusion3m2 attenuator(X+P) | 8.6 | 1.75 | 0.47 | |
| Fusion 1 anti(X+P) | 7.6 | 1.87 | 0.43 |
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 100bp ladder | |
| 2 | Fusion6 anti | X+P |
| 3 | Fusion6 anti | X+P |
| 5 | aptamer12_1M | X+P |
| 6 | aptamer12_1M | X+P |
File:Igku xxbeforexx.xxx
File:Igku xxafterxx.xxx
| Name | quantity | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|---|
| Fusion6 anti(X+P) | 5.0 | 1.76 | 0.20 | |
| aptamer12_1M(X+P) | 3.3 | 2.31 | 0.04 |
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 100bp ladder | |
| 2 | aptamer12_P | X+P |
| 3 | aptamer12_P | X+P |
| 5 | spinach-DT | E+S |
| 6 | spinach-DT | E+S |
File:Igku xxbeforexx.xxx
File:Igku xxafterxx.xxx
| Name | quantity | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|---|
| aptamer12_P(X+P) | 3.4 | 1.72 | 0.26 | |
| spinach-DT(E+S) | 10.1 | 1.57 | 0.47 |
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 100bp ladder | |
| 2 | J23100-RBS-laczα-DT | E+S |
| 3 | J23100-RBS-laczα-DT | E+S |
| 5 | RBS-laczα-DT | E+S |
| 6 | RBS-laczα-DT | E+S |
| 8 | RBS-lysis1-DT | X+P |
| 9 | RBS-lysis1-DT | X+P |
| 11 | RBS-lysis2-DT | X+P |
| 12 | RBS-lysis2-DT | X+P |
File:Igku xxbeforexx.xxx
File:Igku xxafterxx.xxx
| Name | quantity | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|---|
| Pcon-RBS-laczα-DT(E+S) | 5.6 | 2.03 | 0.25 | |
| RBS-laczα-DT(E+S) | 4.4 | 2.04 | 0.33 | |
| RBS-lysis1-DT(X+P) | 5.9 | 2.22 | 0.53 | |
| RBS-lysis2-DT(X+P) | 5.1 | 1.67 | 0.48 |
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 100bp ladder | |
| 2 | RBS-lysis3-DT | X+P |
| 3 | RBS-lysis3-DT | X+P |
| 5 | Pcon-pT181 anti | S+P |
| 6 | Pcon-pT181 anti | S+P |
File:Igku xxbeforexx.xxx
File:Igku xxafterxx.xxx
| Name | quantity | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|---|
| RBS-lysis3-DT(X+P) | 23.4 | 1.72 | 0.88 | |
| Pcon-pT181 anti(S+P) | 31.3 | 1.86 | 0.92 |
Ligation
| state | Vector | Inserter | Ligation High ver.2 | ||
|---|---|---|---|---|---|
| experiment | Pcon-spin-DT(ES) | 10.5µg/mL | pSB6A1(ES) | 8.9µg/mL | |
| experiment | Pcon-spin-DT(ES) | 10.5µg/mL | pSB4K5(ES) | 21.8µg/mL | |
| experiment | Pcon-pT181 anti(S+P) | 31.3µg/mL | spin-DT(X+P) | 10.1µg/mL | |
| experiment | Plac(S+P) | 11.6µg/mL | RBS-lysis3-DT | 23.4µg/mL | |
| experiment | Plux(S+P) | 25µg/mL | RBS-lysis3-DT | 23.4µg/mL | |
| experiment | Ptet-pT181 anti(S+P) | 30.6µg/mL | spin-DT(X+P) | 10.1µg/mL | |
| experiment | Pcon-pT181 attenuator(S+P) | 14.5µg/mL | RBS-laczα-DT(X+P) | 4.4µg/mL | |
PCR
| KaiA(plasmid) | KOD plus | 10x buffer | dNTP | MgSO4 | Primer(661-RBS-KaiA-fwd) | Primer(kaiA-662-rev-ver2) | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 15.5 | 25 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 98°C | 57°C | 68°C | -- |
| 2min | 10s | 30s | 28s | 30cycles |
| KaiB(plasmid) | KOD plus | 10x buffer | dNTP | MgSO4 | Primer(662-RBS-KaiB-fwd-ver2) | Primer(kaiB-663-rev-ver2) | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 15.5 | 25 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 98°C | 57°C | 68°C | -- |
| 2min | 10s | 30s | 28s | 30cycles |
| DT(plasmid) | KOD plus | 10x buffer | dNTP | MgSO4 | Primer(664-DT-fwd-ver2) | Primer(DT-suffix-660-rev-ver2) | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 0.1 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16.4 | 25 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 98°C | 57°C | 68°C | -- |
| 2min | 10s | 30s | 28s | 30cycles |
| P-KaiB(PCR prpduct) | KOD plus | 10x buffer | dNTP | MgSO4 | Primer(prefix-PkaiBC-fwd-ver2) | Primer(PkaiBC-suffix--662-rev-ver2) | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 0.7 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 15.8 | 25 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 98°C | 57°C | 68°C | -- |
| 2min | 10s | 30s | 28s | 30cycles |
| Plac(1)(plasmid) | KOD plus | 10x buffer | dNTP | MgSO4 | Primer(GG0-pSBIC3-fwd-ver2) | Primer(Plac-GG1-rev-ver2) | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 0.13 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16.37 | 25 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 98°C | 57°C | 68°C | -- |
| 2min | 10s | 30s | 60s | 30cycles |
| KaiC(plasmid) | KOD plus | 10x buffer | dNTP | MgSO4 | Primer(GG3-RBS-KaiC-fwd-ver2) | Primer(KaiC-GG4-rev-ver2) | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 15.5 | 25 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 98°C | 57°C | 68°C | -- |
| 2min | 10s | 30s | 60s | 30cycles |
Gel Extraction
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 100bp ladder | |
| 2 | KaiA | |
| 3 | KaiA | |
| 4 | KaiB | |
| 5 | KaiB | |
| 6 | DT | |
| 7 | DT | |
| 8 | P-KaiBC | |
| 9 | P-KaiBC | |
| 10 | nega | |
| 11 | 100bp ladder |
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 100bp ladder | |
| 2 | KaiA | |
| 3 | KaiA | |
| 4 | KaiB | |
| 5 | KaiB | |
| 6 | DT | |
| 7 | DT | |
| 8 | P-KaiBC | |
| 9 | P-KaiBC | |
| 10 | nega | |
| 11 | 100bp ladder |
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | Plac | |
| 2 | Plac | |
| 3 | KaiC | |
| 4 | KaiC | |
| 5 | nega | |
| 6 | 1kb ladder |
 "
"