Team:Carnegie Mellon/KillerRed
From 2013.igem.org
Enpederson (Talk | contribs) |
|||
| (17 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Carnegie_Mellon/Templates/header}} | {{:Team:Carnegie_Mellon/Templates/header}} | ||
| + | <html><br><br> | ||
| + | <center> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/2/2a/Wcarlosw.png" width="154" height="243" alt="Project"> </center> | ||
| + | |||
| + | <h2>KillerRed's structure, chemistry and biophysical properties</h2></html> | ||
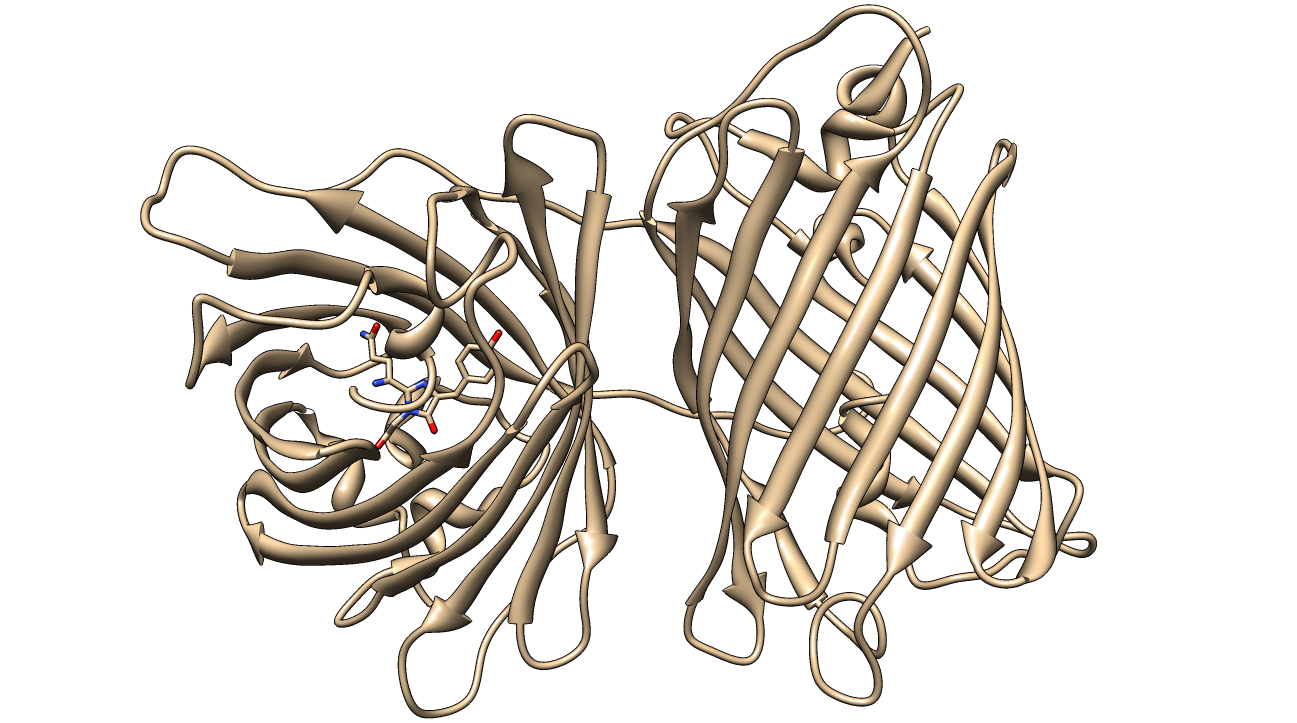
[[image:KillerRed.png|600px|thumb|center|<b>Figure 1: </b>Crystal Structure of KillerRed (PDB ID: 3GB3) with the QYG chromophore shown<sup>2,3,4</sup>]] | [[image:KillerRed.png|600px|thumb|center|<b>Figure 1: </b>Crystal Structure of KillerRed (PDB ID: 3GB3) with the QYG chromophore shown<sup>2,3,4</sup>]] | ||
| - | [[Image:KillerRed BU.jpg|thumb|600px|center|<b>Figure 2: </b>ß-barrel shape of a KillerRed monomer. PDB ID: 2WIQ]] | + | [[Image:KillerRed BU.jpg|thumb|600px|center|<b>Figure 2: </b>ß-barrel shape of a KillerRed monomer.<sup>2,3,4</sup> PDB ID: 2WIQ]] |
<html><br> | <html><br> | ||
<p> | <p> | ||
| - | KillerRed is a red fluorescent protein that produces reactive oxygen species (ROS) in the presence of yellow-orange light (540-585 nm). KillerRed is engineered from anm2CP to be phototoxic. It has been shown that KillerRed produces superoxide radical anions by reacting with water. Superoxide reacts with the chromophore of KillerRed, causing it to become dark, which ultimately gives rise to a bleaching effect. KillerRed is spectrally similar to mRFP1 with a similar brightness. KillerRed is oligomeric and may form large aggregates in cells. Expression of KillerRed and irradiation with light may act a kill-switch for biosafety applications.</p> | + | KillerRed is a red fluorescent protein that produces reactive oxygen species (ROS) in the presence of yellow-orange light (540-585 nm). KillerRed is engineered from anm2CP to be phototoxic. It has been shown that KillerRed produces superoxide radical anions by reacting with water. Superoxide reacts with the chromophore of KillerRed, causing it to become dark, which ultimately gives rise to a bleaching effect. KillerRed is spectrally similar to mRFP1 with a similar brightness. KillerRed is oligomeric and may form large aggregates in cells. Expression of KillerRed and irradiation with light may act as a kill-switch for biosafety applications.</p> |
| + | |||
<p> | <p> | ||
The structure of KillerRed has been analyzed and the structural basis for its toxicity has been explored. The relevant PDB IDs are 2WIQ and 2WIS for the fluorescent and dark states, respectively. </p> | The structure of KillerRed has been analyzed and the structural basis for its toxicity has been explored. The relevant PDB IDs are 2WIQ and 2WIS for the fluorescent and dark states, respectively. </p> | ||
| - | + | <br> | |
<p> | <p> | ||
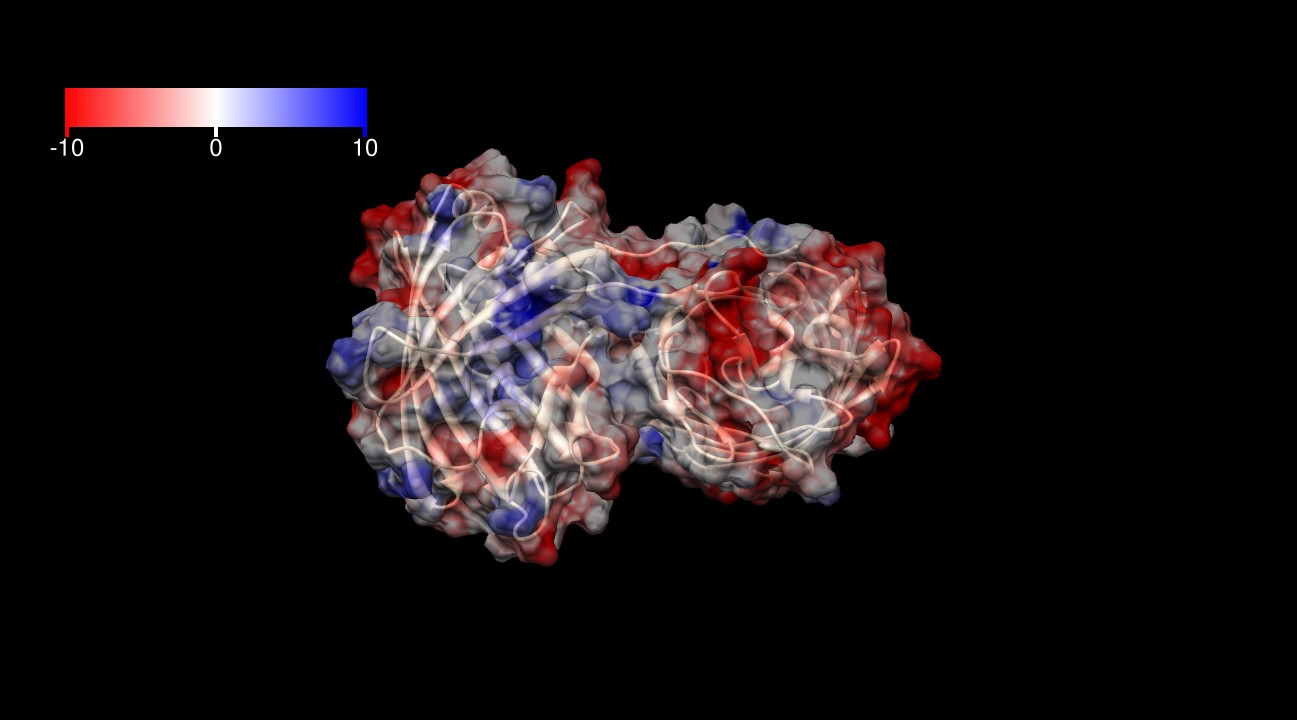
| - | The surface of KillerRed is predominantly negatively charged | + | The surface of KillerRed is predominantly negatively charged (indicated by red areas) as shown in Figures 3-6. The theoretical charge at pH 7.4 is approximately -13 (pI=5.1), which may play a role in its toxicity by interacting more closely with positively charged proteins and positively charged regions of proteins. |
| - | </p/html> | + | </p></html> |
| - | [[Image:KillerRed ES1.jpg|thumb|600px|<b>Figure 3: </b>Electrostatic surface at 1.4 Å of KillerRed dimer (from -10 kcal/(mol*e) in red to +10 kcal(mol*e) in blue). PDB ID: 2WIQ]] | + | [[Image:KillerRed ES1.jpg|thumb|600px|center|<b>Figure 3: </b>Electrostatic surface at 1.4 Å of KillerRed dimer (from -10 kcal/(mol*e) in red to +10 kcal(mol*e) in blue).<sup>3,4</sup> PDB ID: 2WIQ]] |
| - | [[Image:KillerRed ES2.jpg|thumb|600px|<b>Figure 4: </b>Electrostatic surface at 1.4 Å of KillerRed dimer (from -10 kcal/(mol*e) in red to +10 kcal(mol*e) in blue). PDB ID: 2WIQ]] | + | [[Image:KillerRed ES2.jpg|thumb|600px|center|<b>Figure 4: </b>Electrostatic surface at 1.4 Å of KillerRed dimer (from -10 kcal/(mol*e) in red to +10 kcal(mol*e) in blue).<sup>3,4</sup> PDB ID: 2WIQ]] |
| - | [[Image:KillerRed ES3.jpg|thumb|600px|<b>Figure 5: </b>Electrostatic surface at 1.4 Å of KillerRed dimer (from -10 kcal/(mol*e) in red to +10 kcal(mol*e) in blue). PDB ID: 2WIQ]] | + | [[Image:KillerRed ES3.jpg|thumb|600px|center|<b>Figure 5: </b>Electrostatic surface at 1.4 Å of KillerRed dimer (from -10 kcal/(mol*e) in red to +10 kcal(mol*e) in blue).<sup>3,4</sup> PDB ID: 2WIQ]] |
| - | [[Image:KillerRed ES4.jpg|thumb|600px|<b>Figure 6: </b>Electrostatic surface at 1.4 Å of KillerRed dimer (from -10 kcal/(mol*e) in red to +10 kcal(mol*e) in blue). PDB ID: 2WIQ]] | + | [[Image:KillerRed ES4.jpg|thumb|600px|center|<b>Figure 6: </b>Electrostatic surface at 1.4 Å of KillerRed dimer (from -10 kcal/(mol*e) in red to +10 kcal(mol*e) in blue).<sup>3,4</sup> PDB ID: 2WIQ]] |
<html> | <html> | ||
| + | <p> | ||
| + | The interface between the homodimer includes a large network of hydrophobic interactions. This is a phenylalanine- and leucine-rich interface. The H148 residue forms two hydrogen bonds that "connect" the two backbones of the ß-barrels. The histidines and their hydrogen bonds are depicted in Figure 7. | ||
| + | </p> | ||
</html> | </html> | ||
| - | [[Image:KillerRed IntFace.jpg|thumb|600px|<b>Figure 7: </b>The interface between the homodimers consist primarily of hydrophobic interactions and two hydrogen-bonding histidine residues. The histidines and their hydrogen bonds are shown. PDB ID: 2WIQ]] | + | [[Image:KillerRed IntFace.jpg|thumb|600px|center|<b>Figure 7: </b>The interface between the homodimers consist primarily of hydrophobic interactions and two hydrogen-bonding histidine residues. The histidines and their hydrogen bonds are shown. PDB ID: 2WIQ]] |
<html> | <html> | ||
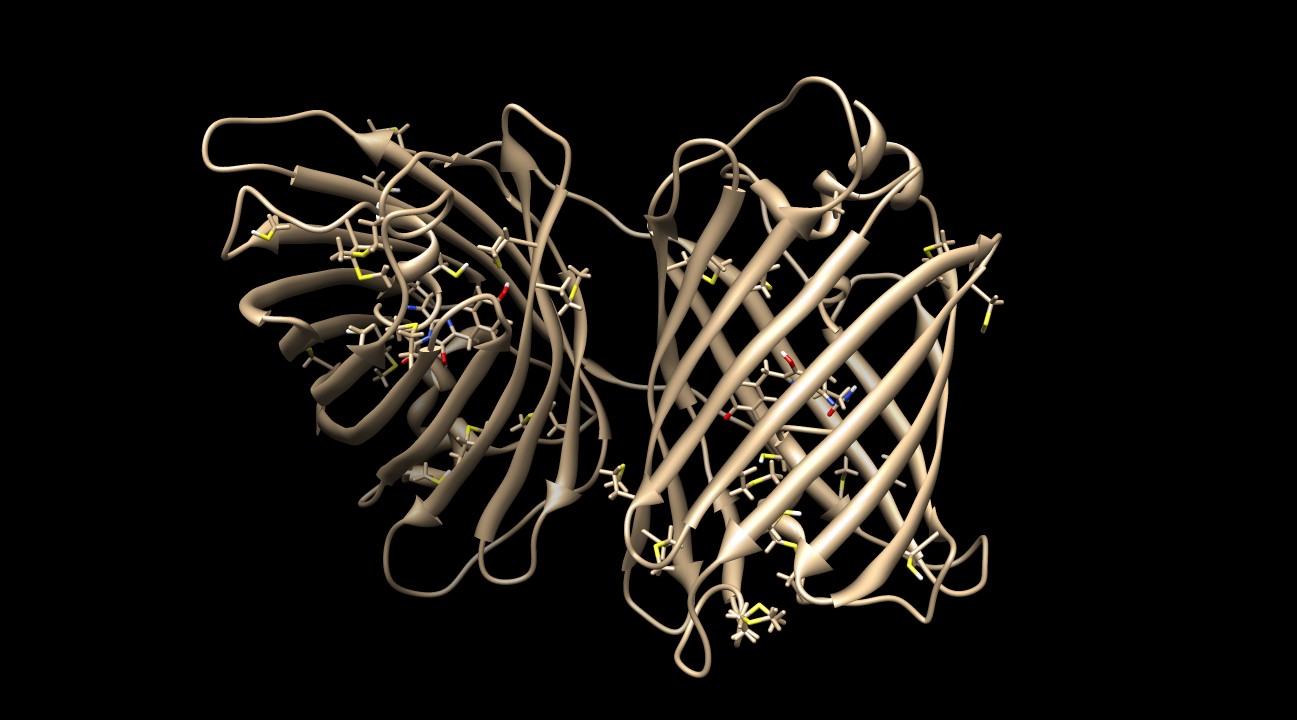
| + | <p> | ||
| + | KillerRed contains many sulfurous residues (10 methionines and 6 cysteines) to protect the protein against ROS damage<sup>2</sup>. Figure 8 depicts the homodimer with these residues visible. | ||
| + | </p> | ||
| + | </html> | ||
| + | [[Image:KillerRed Sulf.jpg|thumb|600px|center|<b>Figure 8: </b>The protein produces superoxide anion radicals, which can react with proteins and lipids to deactivate them. KillerRed contains many sulfurous residues (10 methionines and 6 cysteines) that can act as antioxidants. The large volume of the sulfur atom stabilizes the radical and prevents significant damage to the protein's backbone and side chains.<sup>2,3,4</sup> PDB ID: 2WIQ]] | ||
| + | <html> | ||
| + | <p> | ||
| + | The superoxide anion radical is likely formed in this 8-membered water channel depicted in Figures 10 and 11<sup>2</sup>. The carbonyl on the imidazole ring (Figure 9) hydrogen bonds with the water molecules and reacts with them when the π-electrons in the chromophore are excited. The lifetime of the fluorescence allows enough time for the reaction to occur and the superoxide to be produced<sup>2</sup>. This hypothesis is supported by the fact that fluorescent proteins are light-induced electron donors<sup>6</sup>, which would allow for the electron transfer to the water molecules, causing the superoxide to form. It has been shown that singlet oxygen is also produced by KillerRed, which was originally hypothesized to be the primary ROS produced<sup>1</sup>. More recent data suggests that superoxide is the primary ROS produced by KillerRed<sup>2,7</sup>. It is possible that triplet oxygen can enter the water channel and react in a similar manner to produce singlet oxygen via electron excitation by the chromophore<sup>2</sup>. | ||
| + | </p> | ||
</html> | </html> | ||
| - | [[Image:KillerRed | + | [[Image:CRQ.png|thumb|300px|center|<b>Figure 9: </b> The QYG-derived chromophore of the KillerRed protein ((Z)-2-(2-(4-amino-1-imino-4-oxobutyl)-4-(4-hydroxybenzylidene)-5-oxo-4,5-dihydro-1H-imidazol-1-yl)acetic acid). The extended π-system is highlighted in red. ]] |
| + | [[Image:KillerRed wH.jpg|thumb|600px|center|<b>Figure 10: </b>This is the water channel that produces the superoxide anion radical when exposed to light. The ribbon structure is shown for reference. Na+ is shown in purple.<sup>2,3,4</sup> PDB ID: 2WIQ]] | ||
| + | [[Image:KillerRed WC.jpg|thumb|600px|center|<b>Figure 11: </b>This is the water channel that produces the superoxide anion radical when exposed to light. The hydrogen bonds are shown as pseudobonds and surrounding residues are shown and labeled.<sup>2,3,4</sup>PDB ID: 2WIQ]] | ||
<html> | <html> | ||
| + | <h2>References</h2> | ||
| + | <p><sup>1</sup> Bulina, Maria E, Chudakov, Dmitriy M, Britanova, Olga V, Yanushevich, Yurii G, Staroverov, Dmitry B, Chepurnykh, Tatyana V, Merzlyak, Ekaterina M, Shkrob, Maria A, Lukyanov, Sergey, Lukyanov and Konstantin A. <br /> | ||
| + | <i>A Genetically Encoded Photosensitizer</i><br /> | ||
| + | Nature Biotechnology 2006 24: 95-99. http://dx.doi.org/10.1038/nbt1175 | ||
| + | </p> | ||
| + | <p><sup>2</sup> Sergei Pletnev, Nadya G. Gurskaya, Nadya V. Pletneva, Konstantin A. Lukyanov, Dmitri M. Chudakov, Vladimir I. Martynov, Vladimir O. Popov, Mikhail V. Kovalchuk, Alexander Wlodawer, Zbigniew Dauter, and Vladimir Pletnev.<br /> | ||
| + | <i>Structural Basis for Phototoxicity of the Genetically Encoded Photosensitizer KillerRed</i><br /> | ||
| + | Journal of Biological Chemistry 2009 284: 32028-32039. First Published on September 8, 2009, doi:10.1074/jbc.M109.054973 | ||
| + | </p> | ||
| + | <p><sup>3</sup>Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, funded by grants from the National Institutes of Health National Center for Research Resources (2P41RR001081) and National Institute of General Medical Sciences (9P41GM103311). | ||
| + | </p> | ||
| + | <p><sup>4</sup>Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE.<br /> | ||
| + | <i>UCSF Chimera--a visualization system for exploratory research and analysis.</i><br /> | ||
| + | Journal of Computational Chemistry. 2004 Oct;25(13):1605-12.</p> | ||
| + | <p><sup>5</sup>Takemoto K, Matsuda T, Sakai N, Fu D, Noda M, Uchiyama S, Kotera I, Yoshiyuki A, Horiuchi M, Fukui K, Ayabe T, Inagaki F, Suzuki H, Nagai T. <i>SuperNova, a monomeric photosensitizing fluorescent protein for chromophore-assisted light inactivation</i>. Sci. Rep. 2013/09/17/online. 3. Macmillan Publishers Limited. All rights reserved. http://dx.doi.org/10.1038/srep02629 <br></p> | ||
| + | <p><sup>6</sup>Bogdanov, Alexey M, Mishin, Alexander S, Yampolsky, Ilia V, Belousov, Vsevolod V, Chudakov, Dmitriy M, Subach, Fedor V, Verkhusha, Vladislav V, Lukyanov, Sergey, Lukyanov, Konstantin A. <i>Green fluorescent proteins are light-induced electron donors</i>. Nat Chem Biol. 2009/07//print. 5(7): 459-461. Nature Publishing Group http://dx.doi.org/10.1038/nchembio.174</p> | ||
| + | </p> | ||
| + | <p><sup>7</sup>E.O. Serebrovskaya, E.F. Edelweiss, O.A. Stremovskiy, K.A. Lukyanov, D.M. Chudakov, S.M. Deyev | ||
| + | <i>Targeting cancer cells by using an antireceptor antibody-photosensitizer fusion protein</i> | ||
| + | Proc. Natl. Acad. Sci. USA, 106 (2009), pp. 9221–9225 | ||
| + | </p> | ||
</html> | </html> | ||
| - | |||
| - | |||
Latest revision as of 15:16, 27 September 2013

KillerRed's structure, chemistry and biophysical properties
KillerRed is a red fluorescent protein that produces reactive oxygen species (ROS) in the presence of yellow-orange light (540-585 nm). KillerRed is engineered from anm2CP to be phototoxic. It has been shown that KillerRed produces superoxide radical anions by reacting with water. Superoxide reacts with the chromophore of KillerRed, causing it to become dark, which ultimately gives rise to a bleaching effect. KillerRed is spectrally similar to mRFP1 with a similar brightness. KillerRed is oligomeric and may form large aggregates in cells. Expression of KillerRed and irradiation with light may act as a kill-switch for biosafety applications.
The structure of KillerRed has been analyzed and the structural basis for its toxicity has been explored. The relevant PDB IDs are 2WIQ and 2WIS for the fluorescent and dark states, respectively.
The surface of KillerRed is predominantly negatively charged (indicated by red areas) as shown in Figures 3-6. The theoretical charge at pH 7.4 is approximately -13 (pI=5.1), which may play a role in its toxicity by interacting more closely with positively charged proteins and positively charged regions of proteins.
The interface between the homodimer includes a large network of hydrophobic interactions. This is a phenylalanine- and leucine-rich interface. The H148 residue forms two hydrogen bonds that "connect" the two backbones of the ß-barrels. The histidines and their hydrogen bonds are depicted in Figure 7.
KillerRed contains many sulfurous residues (10 methionines and 6 cysteines) to protect the protein against ROS damage2. Figure 8 depicts the homodimer with these residues visible.
The superoxide anion radical is likely formed in this 8-membered water channel depicted in Figures 10 and 112. The carbonyl on the imidazole ring (Figure 9) hydrogen bonds with the water molecules and reacts with them when the π-electrons in the chromophore are excited. The lifetime of the fluorescence allows enough time for the reaction to occur and the superoxide to be produced2. This hypothesis is supported by the fact that fluorescent proteins are light-induced electron donors6, which would allow for the electron transfer to the water molecules, causing the superoxide to form. It has been shown that singlet oxygen is also produced by KillerRed, which was originally hypothesized to be the primary ROS produced1. More recent data suggests that superoxide is the primary ROS produced by KillerRed2,7. It is possible that triplet oxygen can enter the water channel and react in a similar manner to produce singlet oxygen via electron excitation by the chromophore2.
References
1 Bulina, Maria E, Chudakov, Dmitriy M, Britanova, Olga V, Yanushevich, Yurii G, Staroverov, Dmitry B, Chepurnykh, Tatyana V, Merzlyak, Ekaterina M, Shkrob, Maria A, Lukyanov, Sergey, Lukyanov and Konstantin A.
A Genetically Encoded Photosensitizer
Nature Biotechnology 2006 24: 95-99. http://dx.doi.org/10.1038/nbt1175
2 Sergei Pletnev, Nadya G. Gurskaya, Nadya V. Pletneva, Konstantin A. Lukyanov, Dmitri M. Chudakov, Vladimir I. Martynov, Vladimir O. Popov, Mikhail V. Kovalchuk, Alexander Wlodawer, Zbigniew Dauter, and Vladimir Pletnev.
Structural Basis for Phototoxicity of the Genetically Encoded Photosensitizer KillerRed
Journal of Biological Chemistry 2009 284: 32028-32039. First Published on September 8, 2009, doi:10.1074/jbc.M109.054973
3Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, funded by grants from the National Institutes of Health National Center for Research Resources (2P41RR001081) and National Institute of General Medical Sciences (9P41GM103311).
4Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE.
UCSF Chimera--a visualization system for exploratory research and analysis.
Journal of Computational Chemistry. 2004 Oct;25(13):1605-12.
5Takemoto K, Matsuda T, Sakai N, Fu D, Noda M, Uchiyama S, Kotera I, Yoshiyuki A, Horiuchi M, Fukui K, Ayabe T, Inagaki F, Suzuki H, Nagai T. SuperNova, a monomeric photosensitizing fluorescent protein for chromophore-assisted light inactivation. Sci. Rep. 2013/09/17/online. 3. Macmillan Publishers Limited. All rights reserved. http://dx.doi.org/10.1038/srep02629
6Bogdanov, Alexey M, Mishin, Alexander S, Yampolsky, Ilia V, Belousov, Vsevolod V, Chudakov, Dmitriy M, Subach, Fedor V, Verkhusha, Vladislav V, Lukyanov, Sergey, Lukyanov, Konstantin A. Green fluorescent proteins are light-induced electron donors. Nat Chem Biol. 2009/07//print. 5(7): 459-461. Nature Publishing Group http://dx.doi.org/10.1038/nchembio.174
7E.O. Serebrovskaya, E.F. Edelweiss, O.A. Stremovskiy, K.A. Lukyanov, D.M. Chudakov, S.M. Deyev Targeting cancer cells by using an antireceptor antibody-photosensitizer fusion protein Proc. Natl. Acad. Sci. USA, 106 (2009), pp. 9221–9225
 "
"