Team:Clemson/Notebook
From 2013.igem.org
| Line 13: | Line 13: | ||
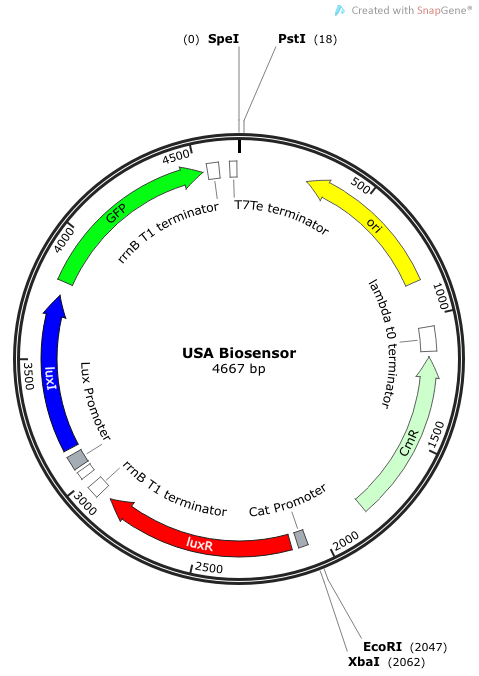
The plasmid construct for the USAB-GFP version is shown below. The other version would replace GFP with amilCP or mAAA. | The plasmid construct for the USAB-GFP version is shown below. The other version would replace GFP with amilCP or mAAA. | ||
<br> | <br> | ||
| + | | | ||
|[[File:USA Biosensor.png|300px|frameless|center]] | |[[File:USA Biosensor.png|300px|frameless|center]] | ||
|[[File:CUTable1.png|400px|frameless|center]] | |[[File:CUTable1.png|400px|frameless|center]] | ||
| + | | | ||
<br> | <br> | ||
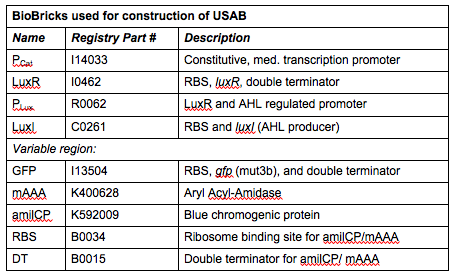
''Rationale'': I14033 was used because the medium transcription rate would be less energetically burdensome on the cell, and this seemed reasonable for a transcriptional regulator. The purpose of having I0462, R0062, and C0261 was to create a positive feedback loop. I0462 would be activated by an outside source of AHLs thereby activating R0062. R0062 would then drive transcription of C0261 to express LuxI, which is responsible for synthesizing more AHLs to drive I0462. This positive feedback loop is what makes the sensor self-amplifying, thus making a very minute signal from a pathogen become more detectable in smaller concentrations. Detection is made possible by GFP or the colorimetric proteins amilCP and mAAA. | ''Rationale'': I14033 was used because the medium transcription rate would be less energetically burdensome on the cell, and this seemed reasonable for a transcriptional regulator. The purpose of having I0462, R0062, and C0261 was to create a positive feedback loop. I0462 would be activated by an outside source of AHLs thereby activating R0062. R0062 would then drive transcription of C0261 to express LuxI, which is responsible for synthesizing more AHLs to drive I0462. This positive feedback loop is what makes the sensor self-amplifying, thus making a very minute signal from a pathogen become more detectable in smaller concentrations. Detection is made possible by GFP or the colorimetric proteins amilCP and mAAA. | ||
Revision as of 22:14, 27 September 2013
Notebook
|
| |||
|
|
 "
"