PET-mCherry
From 2013.igem.org
(→Assembly) |
(→Characterization: wrote intro and developed skeleton) |
||
| Line 30: | Line 30: | ||
==Characterization== | ==Characterization== | ||
| + | The cells for the colonies BEP1 and BEP3 were characterized using flow cytometry. Our goals for this characterization were to show activity of the LacI operator and confirm the expression of mCherry on the outside of the cell. | ||
| + | |||
| + | ===Is the LacI operator working?=== | ||
| + | === Is mCherry on the outside of the cell?=== | ||
| + | |||
==References== | ==References== | ||
<references/> | <references/> | ||
Revision as of 23:04, 27 September 2013
Pet-mCherry is a β-barrel autotransporter with a mCherry passenger.
Contents |
Motivation
In 2012 the Georgia Tech iGEM team developed a novel biosensor based off of green fluorescent protein. The sensor consisted of two subunits of the protein that separately were inactive but once dimerized expressed fluorescence. From this project, we began thinking about how we could develop more complex sensing technology in bacteria. Taking into consideration how mammalian cells sense and react to their environment, we started asking the question: Can bacteria express human integrins?
To start answering this question, we needed to find a way to transport large proteins and anchor them to the outside of the cell. Autodisplay technology seemed like one possible solution to this problem.
Design
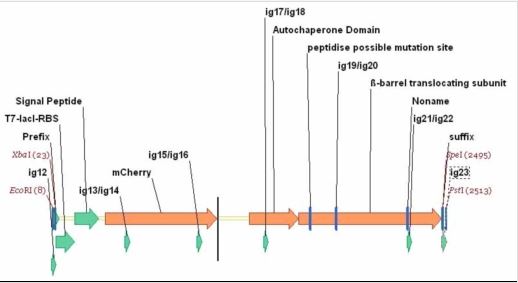
To comply with BioBrick standard 10, Phe-76 was re-optimized to remove a EcoRI site withing the part. A T7 promoter with a lacI operator was added with the RBS (AGGA) to the PET_mCherry. A standard assembly 10 prefix and suffix was then added. In future work, other autodisplay technologies are hoped to be tested to express a number of proteins. [http://www.uniprot.org/uniprot/O68900 Uniprot]
Primers: ig12 SP/Pet_mCherry_b1/0 AGTCAGGAATTCGCGGCCGCTTCTAGAGG ig13 SP/Pet_mCherry_b2/469 CGTATGAAGGCACCCAGACCGCTAAACTGAAA ig14 ASP/Pet_mCherry_b1/500 TTTCAGTTTAGCGGTCTGGGTGCCTTCATACG ig15 SP/Pet_mCherry_b3/928 CGGGTGCTTACAACGTGAACATCAAACTGGAC ig16 ASP/Pet_mCherry_b2/959 GTCCAGTTTGATGTTCACGTTGTAAGCACCCG ig17 SP/Pet_mCherry_b4/1352 CAAACTGGAAGGTGCGAACAACCTGCTGC ig18 ASP/Pet_mCherry_b3/1380 GCAGCAGGTTGTTCGCACCTTCCAGTTTG ig19 SP/Pet_mCherry_b5/1816 TGTTCACCGGTGTTACCATGACCTACACCGAC ig20 ASP/Pet_mCherry_b4/1847 GTCGGTGTAGGTCATGGTAACACCGGTGAACA ig21 SP/Pet_mCherry_b6/2265 GGTTACCAGTTCGACCTGTTCGCTAACGGTGA ig22 ASP/Pet_mCherry_b5/2296 TCACCGTTAGCGAACAGGTCGAACTGGTAACC ig23 ASP/Pet_mCherry_b6/2516 ACTCTGCAGCGGCCGCTACTAGTATTATTATC
Assembly
The 6 "Gblocks" that we synthesized using IDT were assembled using overlap extension PCR.
Characterization
The cells for the colonies BEP1 and BEP3 were characterized using flow cytometry. Our goals for this characterization were to show activity of the LacI operator and confirm the expression of mCherry on the outside of the cell.
Is the LacI operator working?
Is mCherry on the outside of the cell?
References
<references/>
 "
"