Team:Clemson/Parts
From 2013.igem.org
(Prototype team page) |
|||
| (5 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {{Team:Clemson/page-header}} | |
| + | <groupparts>iGEM013 Clemson</groupparts> | ||
| + | [[File:Model Pathogen.png|400px|frameless|center]] | ||
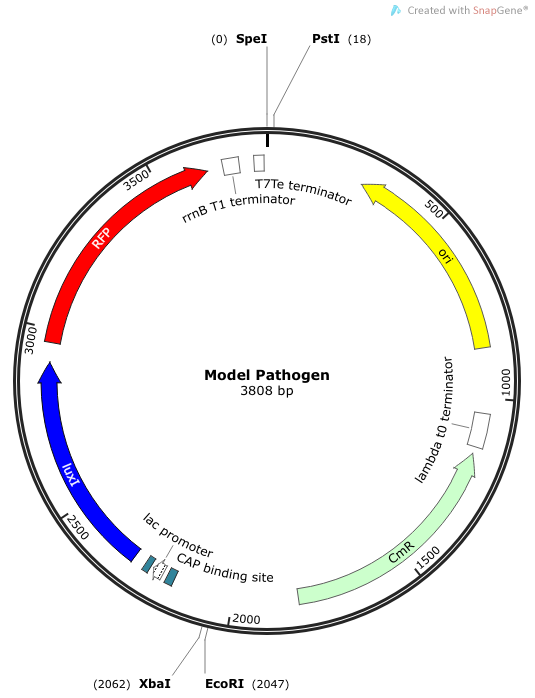
| + | Part BBa_K1090000 was designed to be an AHL signal producer, but RFP is also available as a reporter gene. To quantify the performance of this part, we performed two experiments. The first test the RFP expression in a minimal medium supplemented with lactose or with glucose (as this could affect expression from the lac promoter). The second experiment attempts to indirectly quantify the AHL production by utilizing another construct we made—an AHL signal receiver with GFP signal output. | ||
| + | <br> | ||
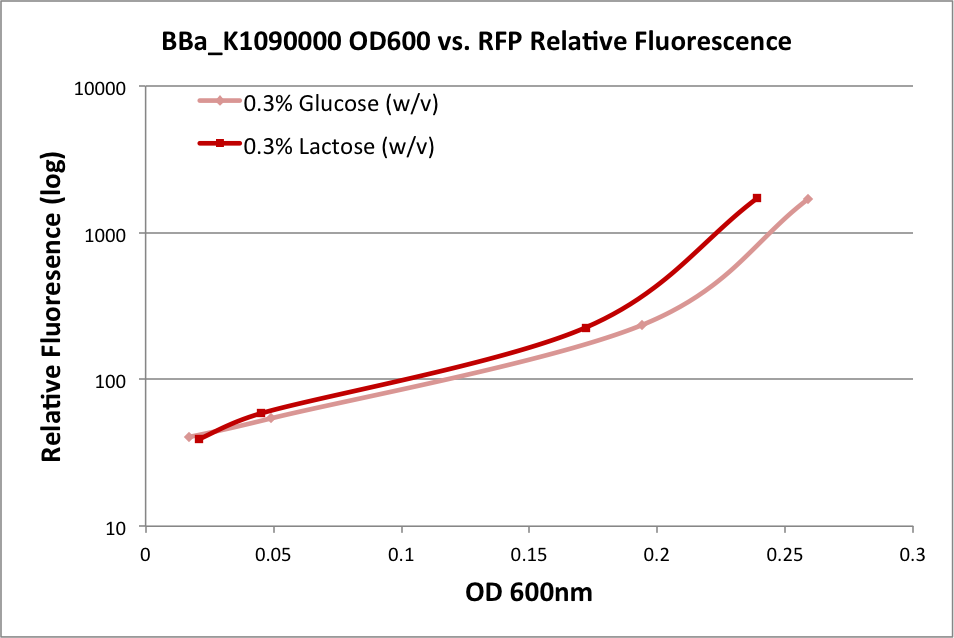
| + | In the first experiment, we grew an overnight culture of E. coli DH10B harboring pSB1C3 with K1090000, harvested and washed the cells, then resuspended them in fresh medium. We used either M9 Miller medium (Miller 1972) supplemented with 0.3% glucose or 0.3% lactose (w/v) with IPTG. (The medium also contained casamino acids and thiamine.) The rate of growth as measured by OD600 of the culture over time indicates it was in early log phase. The OD at 600nm and fluorescence was measured over time, and the relationship is plotted below: | ||
| + | <br> | ||
| + | [[File:main pic 1.png|500px|frameless|center]] | ||
| + | <br> | ||
| + | These results indicate the ability of K1090000 to produce RFP, and indirectly indicate that LuxI and AHL should also be produced. | ||
| + | The second experiment utilized a sensor construct that receives an AHL signal and puts out a GFP signal. Briefly, the construct had luxR (an activator in the presence of AHL), PLux (activated by LuxR + AHL), and GFP. For more info on this construct, please see our 2013 team wiki. | ||
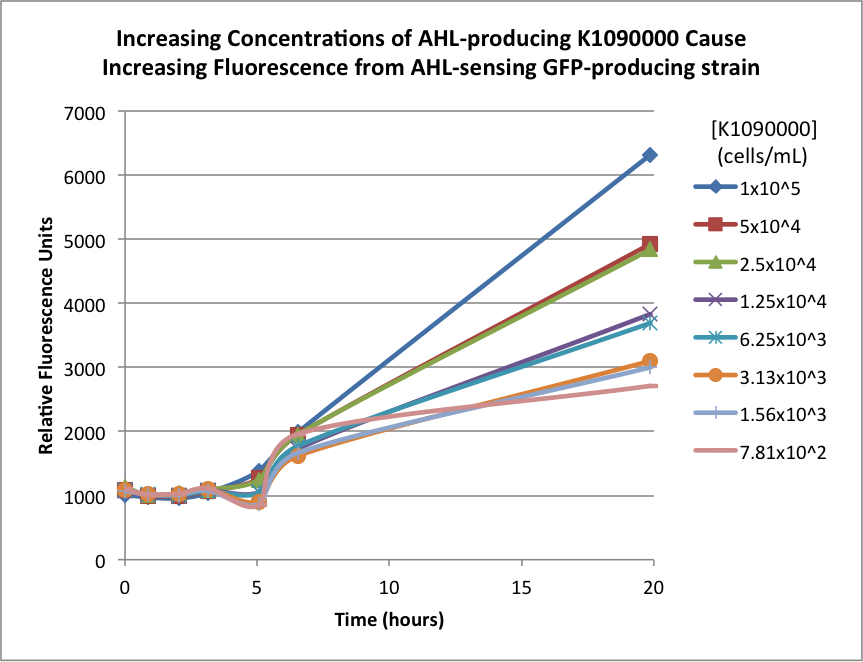
| + | The biosensor and K1090000 were cultured separately overnight, harvested, washed, and resuspended in fresh M9 Miller medium with 0.3% glucose. The biosensor was resuspended to 1x107 cells/mL, and the K1090000 culture was resuspended to 1x105 cells/mL. Serial 50% dilutions of the AHL-producing K1090000 were mixed with the biosensor culture, and the GFP fluorescence was measured over time. The results are shown below: | ||
| + | <br> | ||
| + | [[File:main pic 3.png|500px|frameless|center]] | ||
| + | The higher concentrations of the AHL-producing K1090000 resulted in higher GFP signal. These results seem to indicate the ability of this part to synthesize AHL. | ||
| + | <br> | ||
| + | <br> | ||
| + | References: | ||
| + | 1. Miller, J.H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, 466 pp. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | {{Team:Clemson/page-footer}} | |
| - | + | ||
| - | { | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 23:07, 27 September 2013
Part BBa_K1090000 was designed to be an AHL signal producer, but RFP is also available as a reporter gene. To quantify the performance of this part, we performed two experiments. The first test the RFP expression in a minimal medium supplemented with lactose or with glucose (as this could affect expression from the lac promoter). The second experiment attempts to indirectly quantify the AHL production by utilizing another construct we made—an AHL signal receiver with GFP signal output.
In the first experiment, we grew an overnight culture of E. coli DH10B harboring pSB1C3 with K1090000, harvested and washed the cells, then resuspended them in fresh medium. We used either M9 Miller medium (Miller 1972) supplemented with 0.3% glucose or 0.3% lactose (w/v) with IPTG. (The medium also contained casamino acids and thiamine.) The rate of growth as measured by OD600 of the culture over time indicates it was in early log phase. The OD at 600nm and fluorescence was measured over time, and the relationship is plotted below:
These results indicate the ability of K1090000 to produce RFP, and indirectly indicate that LuxI and AHL should also be produced.
The second experiment utilized a sensor construct that receives an AHL signal and puts out a GFP signal. Briefly, the construct had luxR (an activator in the presence of AHL), PLux (activated by LuxR + AHL), and GFP. For more info on this construct, please see our 2013 team wiki.
The biosensor and K1090000 were cultured separately overnight, harvested, washed, and resuspended in fresh M9 Miller medium with 0.3% glucose. The biosensor was resuspended to 1x107 cells/mL, and the K1090000 culture was resuspended to 1x105 cells/mL. Serial 50% dilutions of the AHL-producing K1090000 were mixed with the biosensor culture, and the GFP fluorescence was measured over time. The results are shown below:
The higher concentrations of the AHL-producing K1090000 resulted in higher GFP signal. These results seem to indicate the ability of this part to synthesize AHL.
References:
1. Miller, J.H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, 466 pp.
 "
"