Team:OUC-China/Model in RNA guardian
From 2013.igem.org
Zhengyuchen (Talk | contribs) |
|||
| Line 229: | Line 229: | ||
<div class="row"> | <div class="row"> | ||
| - | <div class="span9"><p style="font-weight:normal;"><font size="2px">We are trying to establish a model for a tool that can stabilize RNA which consists of 3 parts:<br /><br />1.To reveal the reaction between the RBS in a single mRNA sequence and the GFP coding sequence’s translation initiation rate,we use the RBS calculator to calculate the control group and four experimental groups’ expected fluorescence values without considering the RNase E ‘s effect.By comparing the expected value and the measured value ,we get some necessary parameters for our next step and rule out some unimportant factors.<br /><br />2.We establish ODE equations based on enzyme kinetics and some reasonable assumptions for the interation between the RNase E,mRNA,and Ribosomes to find how Ribosomes inhibit an RNase E cleavage and if we can increase the RNA half-life period theoretically. After we get the experimental data, we will compare our model and the fact, and correct our model so that we can describe the regular patters more precisely and use it in our further work.<br /><br />3.We use some | + | <div class="span9"><p style="font-weight:normal;"><font size="2px">We are trying to establish a model for a tool that can stabilize RNA which consists of 3 parts:<br /><br />1.To reveal the reaction between the RBS in a single mRNA sequence and the GFP coding sequence’s translation initiation rate,we use the RBS calculator to calculate the control group and four experimental groups’ expected fluorescence values without considering the RNase E ‘s effect.By comparing the expected value and the measured value ,we get some necessary parameters for our next step and rule out some unimportant factors.<br /><br />2.We establish ODE equations based on enzyme kinetics and some reasonable assumptions for the interation between the RNase E,mRNA,and Ribosomes to find how Ribosomes inhibit an RNase E cleavage and if we can increase the RNA half-life period theoretically. After we get the experimental data, we will compare our model and the fact, and correct our model so that we can describe the regular patters more precisely and use it in our further work.<br /><br />3.We use some statistical methods to detect the errors in our experiment and just to estimate the accuracy of our experiment.Gillespie algorithm is also use to simulate the tool’s stochastic behavior under noise and find if the experimental data exceed the random error range, if yes, that means the our design indeed stable the mRNA.<br /><a href="https://2013.igem.org/Team:OUC-China/Part I"><img src="https://static.igem.org/mediawiki/2013/a/a8/Ouc-model1.png" height="500" width="600" /></a><br />Protein Expression Preliminary prediction<br /><br /><a href="https://2013.igem.org/Team:OUC-China/Part II"><img src="https://static.igem.org/mediawiki/2013/0/06/Ouc-model2.png" height="500" width="600" /></a><br />Prediction via ODE simulation<br /><br /><a href="https://2013.igem.org/Team:OUC-China/Part III"><img src="https://static.igem.org/mediawiki/2013/4/41/Ouc-model3.png" height="500" width="600" /></a><br />Error Detection And Estimation</font></p> |
Revision as of 23:13, 27 September 2013
Overview
We are trying to establish a model for a tool that can stabilize RNA which consists of 3 parts:
1.To reveal the reaction between the RBS in a single mRNA sequence and the GFP coding sequence’s translation initiation rate,we use the RBS calculator to calculate the control group and four experimental groups’ expected fluorescence values without considering the RNase E ‘s effect.By comparing the expected value and the measured value ,we get some necessary parameters for our next step and rule out some unimportant factors.
2.We establish ODE equations based on enzyme kinetics and some reasonable assumptions for the interation between the RNase E,mRNA,and Ribosomes to find how Ribosomes inhibit an RNase E cleavage and if we can increase the RNA half-life period theoretically. After we get the experimental data, we will compare our model and the fact, and correct our model so that we can describe the regular patters more precisely and use it in our further work.
3.We use some statistical methods to detect the errors in our experiment and just to estimate the accuracy of our experiment.Gillespie algorithm is also use to simulate the tool’s stochastic behavior under noise and find if the experimental data exceed the random error range, if yes, that means the our design indeed stable the mRNA.
Protein Expression Preliminary prediction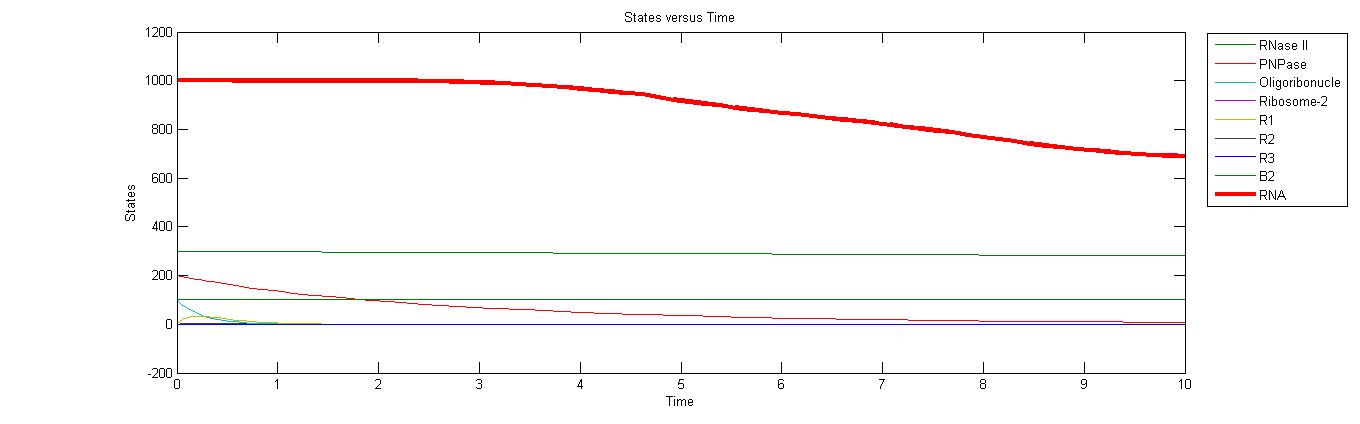
Prediction via ODE simulation
Error Detection And Estimation
 "
"
