Team:BYU Provo/Notebook/Phage Purification/Springexp/Period2/PR
From 2013.igem.org
(Difference between revisions)
| (3 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
{| width="100%" | {| width="100%" | ||
| - | | colspan="3" | <font color="#333399" size="5" font face="Calibri"> ''' | + | | colspan="3" | <font color="#333399" size="5" font face="Calibri"> '''Phage Purification May - June Notebook: May 13 - May 26 Progress Report'''</font> |
<br> | <br> | ||
| Line 10: | Line 10: | ||
|- valign="top" | |- valign="top" | ||
| - | | style="width: | + | | style="width: 20%; background-color: transparent;"| |
<font color="#333399" size="3" font face="Calibri"> | <font color="#333399" size="3" font face="Calibri"> | ||
| - | : | + | <font size = "4"> |
| + | |||
| + | : <u> '''Phage Purification''' </u> </font> | ||
: [[Team:BYU Provo/Notebook/Phage_Purification/Winterexp|March-April]] | : [[Team:BYU Provo/Notebook/Phage_Purification/Winterexp|March-April]] | ||
| Line 26: | Line 28: | ||
</font> | </font> | ||
| - | | style="width: | + | | style="width: 80%; background-color: transparent;"| |
<font face="Calibri" size="3"> | <font face="Calibri" size="3"> | ||
| Line 32: | Line 34: | ||
'''1. Goals for the week ''' | '''1. Goals for the week ''' | ||
| - | : Our | + | : Our goals for this week are to correctly prepare and successfully extract phage from a CsCl gradient. |
'''2.Experiments and results ''' | '''2.Experiments and results ''' | ||
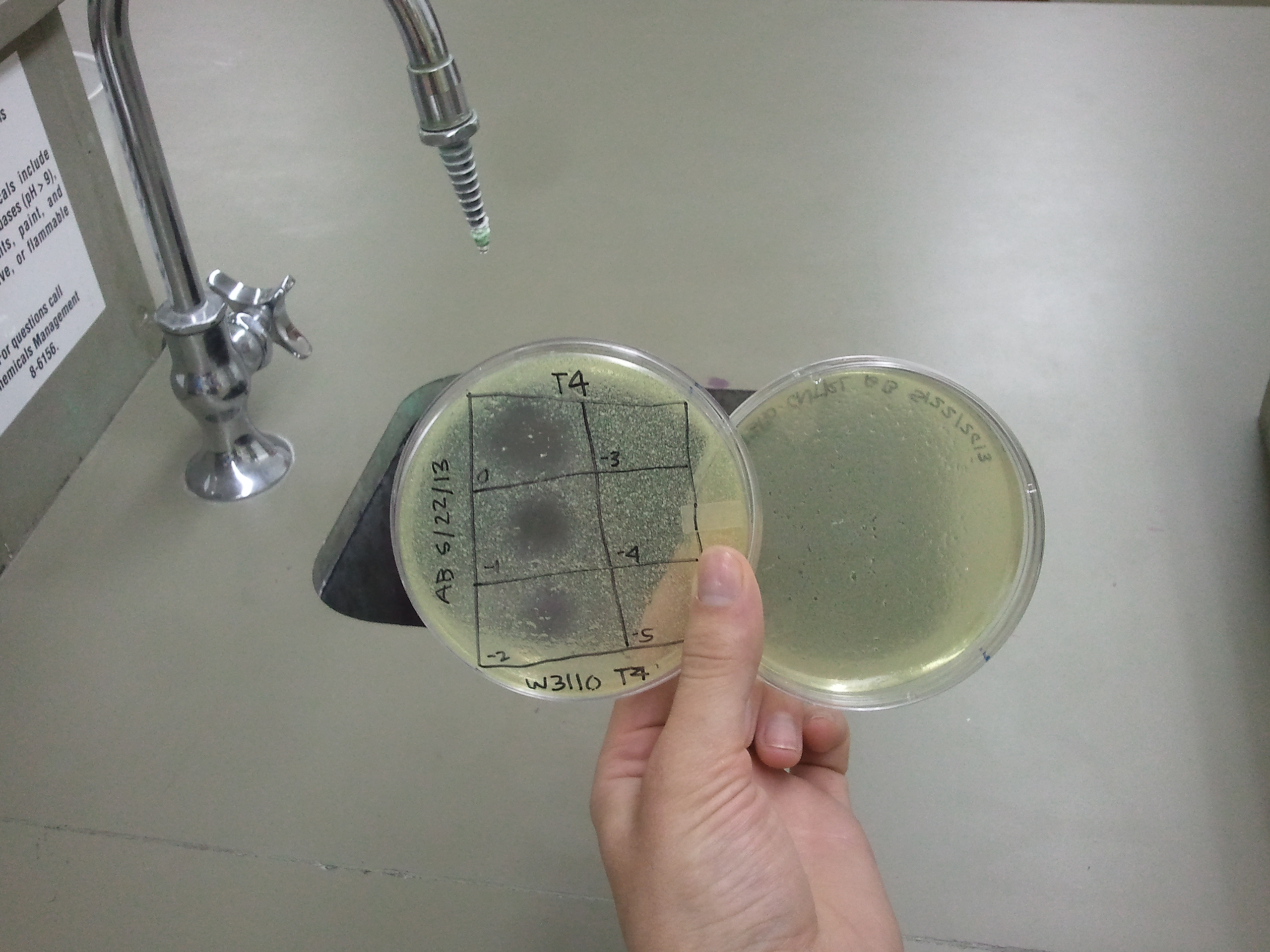
| - | + | :After our first attempt at purification using PEG, we decided to perform a phage titer to check for phage viability. The titers were successful. Both plates showed plaques. On the plate of W3110 infected with T4 phage, there was contamination. Plaques were still visible and it was determined that the team should continue on, as the next step of a CsCl gradient should remove any contaminants. | |
| - | : | + | [[File:w3110_titer_5.22.13.png | thumb|none|alt=A phage plaques were found but with contamination.|w3110]] |
| + | [[File:BL21_5.22.13.png | thumb|none|alt=A phage plaques were found, but they had run.|BL21]] | ||
| + | :When performing the next step of the CsCl gradient, we added the incorrect amount of CsCl to phage suspension buffer, causing the gradient to be half as dense as it should have been. We had layered in very little phage to begin with, and the team determined it would be better to restart with a larger amount of phage particles to purify. | ||
| + | :We completed the first step in PEG purification for the second time. With the doubled amount of phage, we had much more to layer in the CsCl gradient and there were visible bands in the gradient since the correct concentrations were used. | ||
| - | + | :The CsCl gradient was successful the second time it was ran. There were visible bands of bacterial debris and phage particles. | |
| - | + | '''3. Next Steps''' | |
| - | : | + | :Successfully extract phage particles from the CsCl gradient, remove from CsCl using phage suspension buffer. |
| + | : Perform the third and final step of purification with an equilibrium CsCl gradient. | ||
| - | + | :BeginPreparing for the preperation of Ghost Particles. | |
| - | + | ||
| - | + | ||
| - | : | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
Latest revision as of 00:11, 28 September 2013
| Phage Purification May - June Notebook: May 13 - May 26 Progress Report
| ||
|
|
1. Goals for the week
3. Next Steps
| |
 "
"