Team:GeorgiaTech/GFP KQAGDV
From 2013.igem.org
Jackjenkins (Talk | contribs) (→Background) |
Jackjenkins (Talk | contribs) |
||
| (2 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{:Team:GeorgiaTech/template}} | ||
== Background == | == Background == | ||
Green Fluorescent Protein, often referred to as GFP, is a well-known and highly documented protein naturally found in Aequoria victoria (a jellyfish), but has been isolated and optimized for use in many other organisms such as ours in Escherichia coli. GFP is commonly used to easily indicate expression of a gene or a certain binding that is being studied. | Green Fluorescent Protein, often referred to as GFP, is a well-known and highly documented protein naturally found in Aequoria victoria (a jellyfish), but has been isolated and optimized for use in many other organisms such as ours in Escherichia coli. GFP is commonly used to easily indicate expression of a gene or a certain binding that is being studied. | ||
| - | + | ||
Our interest with this part began with our interest in integrins, which are the receptors present on mammalian cells that allow communication across the extracellular matrix. | Our interest with this part began with our interest in integrins, which are the receptors present on mammalian cells that allow communication across the extracellular matrix. | ||
| Line 11: | Line 12: | ||
First, we designed in silica our construct, with a known GFP vector and inserting the KQAGDV at the end of this sequence. | First, we designed in silica our construct, with a known GFP vector and inserting the KQAGDV at the end of this sequence. | ||
[[File:GFP_KQAGDV.png]] | [[File:GFP_KQAGDV.png]] | ||
| - | + | ||
To construct this, three primers were designed to perform an overlap extension PCR reaction, a process that allows certain primers to recognize DNA within the vector, match it and expand upon it: | To construct this, three primers were designed to perform an overlap extension PCR reaction, a process that allows certain primers to recognize DNA within the vector, match it and expand upon it: | ||
[[File:Primers1.png]] | [[File:Primers1.png]] | ||
| - | + | ||
Once this successfully added the KQAGDV tag, we were able to transform this vector, purify it, and send it for sequencing. | Once this successfully added the KQAGDV tag, we were able to transform this vector, purify it, and send it for sequencing. | ||
| Line 23: | Line 24: | ||
[[File:Primers2.png]] | [[File:Primers2.png]] | ||
| - | + | ||
Using these primers, we set up a Site-Directed Mutagenesis reaction (SDM) which is very effective for inserting or fixing a small number of base pairs within a sequence. This reaction grew successful colonies, which were then purified and sent for sequencing. | Using these primers, we set up a Site-Directed Mutagenesis reaction (SDM) which is very effective for inserting or fixing a small number of base pairs within a sequence. This reaction grew successful colonies, which were then purified and sent for sequencing. | ||
| - | + | ||
The results from this sequencing had some issues; the EcoR1 site was mutated and the promoter region was lost from the gene. | The results from this sequencing had some issues; the EcoR1 site was mutated and the promoter region was lost from the gene. | ||
| - | + | ||
While trying to determine the best way to fix this, we moved on with the project by inserting a 6HIS site in front of the GFP. Polyhistedene tags function as a detector for protein-protein interactions, and will help us detect that our GFP protein is indeed interacting with the αIIbβ3 integrin structure. | While trying to determine the best way to fix this, we moved on with the project by inserting a 6HIS site in front of the GFP. Polyhistedene tags function as a detector for protein-protein interactions, and will help us detect that our GFP protein is indeed interacting with the αIIbβ3 integrin structure. | ||
| - | + | ||
This process was achieved through an overlap extension PCR with the following primers: | This process was achieved through an overlap extension PCR with the following primers: | ||
[[File:Primers3.png]] | [[File:Primers3.png]] | ||
| - | + | ||
We successfully added the 6HIS site, but the previous problems (lack of promoter, damaged EcoR1 site) were still present. This was fixed with a couple of steps. The first was to amplify the construct using the BioBrick primers to format this vector into the BioBrick format. Once this was done, the vector was cut at the Xba and Pst sites and combined with a BioBrick we created that had a T7 promoter-lacI operator gene and these were ligated together using 3A Assembly method. | We successfully added the 6HIS site, but the previous problems (lack of promoter, damaged EcoR1 site) were still present. This was fixed with a couple of steps. The first was to amplify the construct using the BioBrick primers to format this vector into the BioBrick format. Once this was done, the vector was cut at the Xba and Pst sites and combined with a BioBrick we created that had a T7 promoter-lacI operator gene and these were ligated together using 3A Assembly method. | ||
| - | |||
==Future Implications== | ==Future Implications== | ||
With a few small fixes, this construct will be usable for its purpose in binding to αIIbβ3 integrin subunits and fluoresce when this binding occurs. It is necessary to fix the EcoRI restriction site and add it to a chloramphenicol backbone to successfully place it in a BioBrick standard. For it to be succesfully used and indicate when the binding occurs, the promoter region needs to be re-inserted into the vector. | With a few small fixes, this construct will be usable for its purpose in binding to αIIbβ3 integrin subunits and fluoresce when this binding occurs. It is necessary to fix the EcoRI restriction site and add it to a chloramphenicol backbone to successfully place it in a BioBrick standard. For it to be succesfully used and indicate when the binding occurs, the promoter region needs to be re-inserted into the vector. | ||
Latest revision as of 02:10, 28 September 2013

Main Page

Background
Green Fluorescent Protein, often referred to as GFP, is a well-known and highly documented protein naturally found in Aequoria victoria (a jellyfish), but has been isolated and optimized for use in many other organisms such as ours in Escherichia coli. GFP is commonly used to easily indicate expression of a gene or a certain binding that is being studied.
Our interest with this part began with our interest in integrins, which are the receptors present on mammalian cells that allow communication across the extracellular matrix.
In order to readily and systematically detect active integrins present on specific mammalian cells of interest, we designed a GFP-KQAGDV sequence, which is a vector containing a green fluorescent protein with a KQAGDV tag. This tag acts as a ligand that binds with the αIIbβ3, an integrin pairing common on mammalian red blood cells. Binding to this integrin would leave a clear marker with the GFP to allow the integrin to be seen using flow cytometry.
Project
First, we designed in silica our construct, with a known GFP vector and inserting the KQAGDV at the end of this sequence.
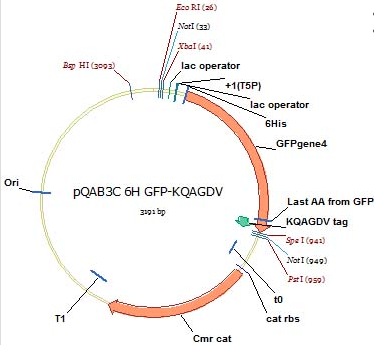
To construct this, three primers were designed to perform an overlap extension PCR reaction, a process that allows certain primers to recognize DNA within the vector, match it and expand upon it:
Once this successfully added the KQAGDV tag, we were able to transform this vector, purify it, and send it for sequencing.
Sequencing results showed us that there was a deletion of an AGGTGA sequence within the KQAGDV site. Primers were designed and ordered to fix this:
Using these primers, we set up a Site-Directed Mutagenesis reaction (SDM) which is very effective for inserting or fixing a small number of base pairs within a sequence. This reaction grew successful colonies, which were then purified and sent for sequencing.
The results from this sequencing had some issues; the EcoR1 site was mutated and the promoter region was lost from the gene.
While trying to determine the best way to fix this, we moved on with the project by inserting a 6HIS site in front of the GFP. Polyhistedene tags function as a detector for protein-protein interactions, and will help us detect that our GFP protein is indeed interacting with the αIIbβ3 integrin structure.
This process was achieved through an overlap extension PCR with the following primers:
![]()
We successfully added the 6HIS site, but the previous problems (lack of promoter, damaged EcoR1 site) were still present. This was fixed with a couple of steps. The first was to amplify the construct using the BioBrick primers to format this vector into the BioBrick format. Once this was done, the vector was cut at the Xba and Pst sites and combined with a BioBrick we created that had a T7 promoter-lacI operator gene and these were ligated together using 3A Assembly method.
Future Implications
With a few small fixes, this construct will be usable for its purpose in binding to αIIbβ3 integrin subunits and fluoresce when this binding occurs. It is necessary to fix the EcoRI restriction site and add it to a chloramphenicol backbone to successfully place it in a BioBrick standard. For it to be succesfully used and indicate when the binding occurs, the promoter region needs to be re-inserted into the vector.
 "
"
