Team:Carnegie Mellon/Project/Results
From 2013.igem.org
Enpederson (Talk | contribs) |
Enpederson (Talk | contribs) |
||
| (13 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Carnegie_Mellon/Templates/header}} | {{:Team:Carnegie_Mellon/Templates/header}} | ||
| - | <br> | + | <br><br><br> |
| - | [[ | + | <h2>KillerRed phototoxicity</h2> |
| + | Our photobleaching experiments showed the phototoxicity of KillerRed in pSB1A3:<br> | ||
| + | |||
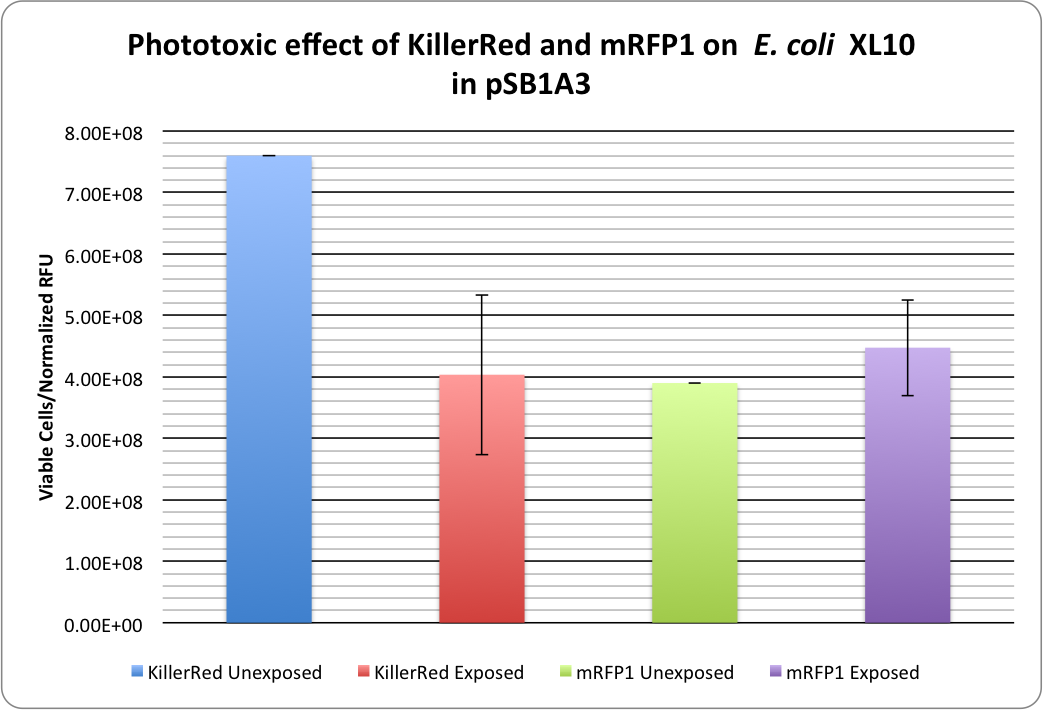
| + | [[image:KillerRed RFP Phototoxicity1.png|thumb|400px|center|<b>Figure 1:</b> Phototoxicity of KillerRed and mRFP1 (<partinfo>BBa_E1010</partinfo>) on <i>E. coli</i> XL10 by irradiating with a HBO100 lamp (includes 375 nm LP filter) for 5 hours.]] | ||
| + | <br><br> | ||
<p>KillerRed's phototoxic effect on <i>E. coli</i> XL10 is shown in Figure 1.<br /> | <p>KillerRed's phototoxic effect on <i>E. coli</i> XL10 is shown in Figure 1.<br /> | ||
| + | <br> | ||
| + | Triplicate cell viabilities are divided by the normalized fluorescence values to account for bleaching effects and the fact that KillerRed has a lower expression level than mRFP1. | ||
RFP 114% ± 20% (viable cells)/(Normalized RFU) <br /> | RFP 114% ± 20% (viable cells)/(Normalized RFU) <br /> | ||
KillerRed: 53% ± 17% (viable cells)/(Normalized RFU) | KillerRed: 53% ± 17% (viable cells)/(Normalized RFU) | ||
| - | </p> | + | </p><br><br> |
| - | [[ | + | <h2>KillerRed photobleaching curve</h2> |
| + | |||
| + | [[image:KR-Photobleach.png|thumb|400px|center|<b>Figure 1:</b> Photobleaching curve of KillerRed with a HBO100 mercury-arc lamp]] | ||
| + | <br><br> | ||
<p>XL10 Ultracompetent cells were transformed with KillerRed (BBa_K1184000) cloned with <partinfo>BBa_B0034</partinfo> as the RBS and <partinfo>BBa_R0010</partinfo> as the wild-type lac promoter and induced overnight with IPTG.The overnight was bleached for 180 minutes with HBO100 (100W Mercury-arc lamp). Fluorescence data was taken using a Tecan Safire II with the parameters shown in Table 1. Fluorescence Data is shown in Table 2.</p> | <p>XL10 Ultracompetent cells were transformed with KillerRed (BBa_K1184000) cloned with <partinfo>BBa_B0034</partinfo> as the RBS and <partinfo>BBa_R0010</partinfo> as the wild-type lac promoter and induced overnight with IPTG.The overnight was bleached for 180 minutes with HBO100 (100W Mercury-arc lamp). Fluorescence data was taken using a Tecan Safire II with the parameters shown in Table 1. Fluorescence Data is shown in Table 2.</p> | ||
| - | <table cellpadding="2" border="1px" cellspacing="0" align="center" width=" | + | <table cellpadding="2" border="1px" cellspacing="0" align="center" width="40%"> |
| + | <br> | ||
<caption><p align="justify"><b>Table 1</b> Tecan Safire<sup>2</sup> Parameters</p></caption> | <caption><p align="justify"><b>Table 1</b> Tecan Safire<sup>2</sup> Parameters</p></caption> | ||
<tr><td><b>Excitation (nm)</b></td><td>585</td></tr> | <tr><td><b>Excitation (nm)</b></td><td>585</td></tr> | ||
| Line 17: | Line 27: | ||
<tr><td><b>Number of reads</b></td><td>10</td></tr> | <tr><td><b>Number of reads</b></td><td>10</td></tr> | ||
<tr><td><b>Integration Time (microseconds)</b></td><td>40</td></tr> | <tr><td><b>Integration Time (microseconds)</b></td><td>40</td></tr> | ||
| + | <br><br><br> | ||
</table> | </table> | ||
| - | <table cellpadding="2" border="1px" cellspacing="0" align="center" width=" | + | <table cellpadding="2" border="1px" cellspacing="0" align="center" width="40%"> |
<caption><p align="justify"><b>Table 2</b> Shows the fluorescence data over time during photobleaching.</p></caption> | <caption><p align="justify"><b>Table 2</b> Shows the fluorescence data over time during photobleaching.</p></caption> | ||
<tr><td align="center"><b>Time (minutes)</b></td><td align="center"><b>Fluorescence (RFU)</b></td> | <tr><td align="center"><b>Time (minutes)</b></td><td align="center"><b>Fluorescence (RFU)</b></td> | ||
| Line 32: | Line 43: | ||
<tr><td align="left">180</td><td align="right">13741</td></tr> | <tr><td align="left">180</td><td align="right">13741</td></tr> | ||
</table> | </table> | ||
| + | <p><html> | ||
| + | To read our conclusions and future directions, visit our <a href="2013.igem.org/Team:Carnegie_Mellon/Project/Abstract">Abstract</a> and <a href="https://2013.igem.org/Team:Carnegie_Mellon/Project/Future">Future Directions</a> pages. | ||
| + | <br><br></html> | ||
Latest revision as of 03:39, 28 September 2013
KillerRed phototoxicity
Our photobleaching experiments showed the phototoxicity of KillerRed in pSB1A3:
KillerRed's phototoxic effect on E. coli XL10 is shown in Figure 1.
Triplicate cell viabilities are divided by the normalized fluorescence values to account for bleaching effects and the fact that KillerRed has a lower expression level than mRFP1.
RFP 114% ± 20% (viable cells)/(Normalized RFU)
KillerRed: 53% ± 17% (viable cells)/(Normalized RFU)
KillerRed photobleaching curve
XL10 Ultracompetent cells were transformed with KillerRed (BBa_K1184000) cloned with <partinfo>BBa_B0034</partinfo> as the RBS and <partinfo>BBa_R0010</partinfo> as the wild-type lac promoter and induced overnight with IPTG.The overnight was bleached for 180 minutes with HBO100 (100W Mercury-arc lamp). Fluorescence data was taken using a Tecan Safire II with the parameters shown in Table 1. Fluorescence Data is shown in Table 2.
| Excitation (nm) | 585 |
| Emission (nm) | 610 |
| Excitation bandwidth (nm) | 10 |
| Emission bandwidth (nm) | 10 |
| Gain | 129 |
| Number of reads | 10 |
| Integration Time (microseconds) | 40 |
| Time (minutes) | Fluorescence (RFU) |
| 0 | 42598 |
| 20 | 37616 |
| 40 | 33749 |
| 60 | 29059 |
| 80 | 25680 |
| 100 | 21985 |
| 120 | 19442 |
| 140 | 17031 |
| 160 | 15738 |
| 180 | 13741 |
To read our conclusions and future directions, visit our Abstract and Future Directions pages.
 "
"