Team:DTU-Denmark/Notebook/6 August 2013
From 2013.igem.org
(→plasmid miniprep) |
|||
| (31 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:DTU-Denmark/Templates/StartPage|6 August 2013}} | {{:Team:DTU-Denmark/Templates/StartPage|6 August 2013}} | ||
| - | + | Navigate to the [[Team:DTU-Denmark/Notebook/5_August_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/7_August_2013|Next]] Entry | |
| - | = | + | =Lab 208= |
<hr/> | <hr/> | ||
==Main purpose== | ==Main purpose== | ||
<hr/> | <hr/> | ||
| + | * Extraction PCR for AMO | ||
| + | * PCR for SPL (synthetic promoter library) | ||
| + | * PCR for reference promoter | ||
| + | * PCR for araBAD biobrick K808000 | ||
| + | * PCR to extract Nir2 from ''P. aeruginosa'' | ||
| + | * PCR for araBAD and SPL with DMSO | ||
| + | * PCR on Nir1 with MgCl2 gradient | ||
| + | * Gel purification of Nir1 (from ''P. aeruginosa'') | ||
| + | * Plasmid isolation: HAO in pZA21 from USER cloning. | ||
==Who was in the lab== | ==Who was in the lab== | ||
| Line 13: | Line 22: | ||
<hr/> | <hr/> | ||
| - | + | ===PCR for AMO=== | |
Made four reactions using 1 uL respectively 10 uL of culture from the -20 or from the glycerol stock (-80) as template and adjusted volume of water accordingly. | Made four reactions using 1 uL respectively 10 uL of culture from the -20 or from the glycerol stock (-80) as template and adjusted volume of water accordingly. | ||
| Line 19: | Line 28: | ||
Primers 10a, 10b. | Primers 10a, 10b. | ||
| - | + | ===PCR for SPL (synthetic promoter library)=== | |
| - | template: | + | template: pZA21 with RFP, primers: 52a, 52b1, temperature: 50C, time: 2:30 |
| - | + | ===PCR for reference promoter=== | |
| - | template: | + | template: pZA21 with RFP, primers: 52a, 52b2, temperature: 50C, time: 2:30 |
| - | + | ===PCR for araBAD K808000=== | |
template: K808000, primers: 12a, 12bn, temperature: 50C, time: 2:30 | template: K808000, primers: 12a, 12bn, temperature: 50C, time: 2:30 | ||
| - | === | + | ===PCR to extract Nir2 from P. aeruginosa=== |
| - | + | ===PCR for araBAD and SPL with DMSO=== | |
| + | Template and primers as previous. | ||
| - | === | + | ===PCR on Nir1 with MgCl2 gradient=== |
| + | Used previous isolation from a gel purification to make PCR for amplification of the strand. Used 5% DMSO as additive. The PCR was done with primer pair 41 and with x7 polymerase at 55C annealing and 5:00 min extension. Also we used 20 sec as denaturing time instead of the normal 10. | ||
| + | The magnesium gradient was as follow: | ||
| + | *0 | ||
| + | *1uL MgCl2 2mM per 50uL reaction | ||
| + | *5uL MgCl2 2mM per 50uL reaction | ||
| + | *20uL MgCl2 2mM per 50uL reaction | ||
| + | |||
| + | ===Gel purification=== | ||
| + | |||
| + | of the Nir1 fragment extracted from ''P. aeruginosa''. Nanodrop: 8ng/uL | ||
| + | |||
| + | ===Plasmid miniprep=== | ||
HAO in pZA21 from USER cloning was purified for restriction analysis. Nanodrop measurement: 180ng/uL | HAO in pZA21 from USER cloning was purified for restriction analysis. Nanodrop measurement: 180ng/uL | ||
| Line 42: | Line 64: | ||
<hr/> | <hr/> | ||
| + | ===Gel 1=== | ||
1% agarose gel | 1% agarose gel | ||
* 1 kb ladder | * 1 kb ladder | ||
| - | * Nir1 col. | + | * Nir1 col. Taq |
| - | * Nir1 col. | + | * Nir1 col. Taq |
| - | * Nir2 col. | + | * Nir2 col. Taq |
| - | * Nir2 col. | + | * Nir2 col. Taq |
| - | * Nir1 frag. | + | * Nir1 frag. Taq |
| - | * Nir1 frag. | + | * Nir1 frag. Taq |
| - | * Nir1 col. | + | * Nir1 col. x7 |
| - | * Nir1 col. | + | * Nir1 col. x7 |
* neg | * neg | ||
| - | * Hi | + | * Hi spec. buffer |
| - | * Hi | + | * Hi spec. buffer |
| - | * DMSO | + | * DMSO |
* DMSO | * DMSO | ||
* Nir1 | * Nir1 | ||
| Line 66: | Line 89: | ||
[[File:2013-08-06 nir.jpg| 600px]] | [[File:2013-08-06 nir.jpg| 600px]] | ||
| + | Nir1 was purified. | ||
| + | |||
| + | ===Gel 2=== | ||
1% agarose gel | 1% agarose gel | ||
| Line 81: | Line 107: | ||
* 1 kb ladder | * 1 kb ladder | ||
| - | + | [[File:2013-08-06 small gel.jpg|600px]] | |
| - | ===Gel | + | ===Gel 3=== |
| - | * 1 | + | 1% agarose gel |
| - | * | + | |
| - | * | + | * 1 kb ladder |
| - | * | + | * AMO, 1uL of -20 culture as template |
| - | * | + | * AMO, 10uL of -20 culture as template |
| - | * | + | * AMO, 1uL of glycerol stock as template |
| - | * | + | * AMO, 10uL of glycerol stock as template |
| - | * | + | * neg from AMO PCR |
| - | + | * 1 - SPL, primers 52a, 52b1, template pZA21 with RFP, 50C, 2:30 min extension | |
| - | + | * 2 - SPL, primers 52a, 52b1, template pZA21 with RFP, 50C, 2:30 min extension | |
| - | + | * 3 - ref, primers 52a, 52b2, template pZA21 with RFP, 50C, 2:30 min extension | |
| - | + | * 4 - ref, primers 52a, 52b2, template pZA21 with RFP, 50C, 2:30 min extension | |
| - | + | * 5 - araBAD, primers 12a, 12bn, template K80800, 50C, 2:30 min extension | |
| - | + | * 6 - araBAD, primers 12a, 12bn, template K80800, 50C, 2:30 min extension | |
| - | + | * 1 kb ladder | |
| - | + | ||
| - | + | [[File:2013-08-06 AMO and spl.jpg| 600px]] | |
| - | + | ||
| + | AMO was gel purified. | ||
==Conclusion== | ==Conclusion== | ||
<hr/> | <hr/> | ||
| + | *Nir was obtained from colony PCR and purified. | ||
| + | *AMO was extracted and gel purified. | ||
| + | |||
Navigate to the [[Team:DTU-Denmark/Notebook/5_August_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/7_August_2013|Next]] Entry | Navigate to the [[Team:DTU-Denmark/Notebook/5_August_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/7_August_2013|Next]] Entry | ||
{{:Team:DTU-Denmark/Templates/EndPage}} | {{:Team:DTU-Denmark/Templates/EndPage}} | ||
Latest revision as of 22:09, 29 September 2013
6 August 2013
Contents |
Lab 208
Main purpose
- Extraction PCR for AMO
- PCR for SPL (synthetic promoter library)
- PCR for reference promoter
- PCR for araBAD biobrick K808000
- PCR to extract Nir2 from P. aeruginosa
- PCR for araBAD and SPL with DMSO
- PCR on Nir1 with MgCl2 gradient
- Gel purification of Nir1 (from P. aeruginosa)
- Plasmid isolation: HAO in pZA21 from USER cloning.
Who was in the lab
Kristian, Gosia, Natalia, Henrike
Procedure
PCR for AMO
Made four reactions using 1 uL respectively 10 uL of culture from the -20 or from the glycerol stock (-80) as template and adjusted volume of water accordingly.
Primers 10a, 10b.
PCR for SPL (synthetic promoter library)
template: pZA21 with RFP, primers: 52a, 52b1, temperature: 50C, time: 2:30
PCR for reference promoter
template: pZA21 with RFP, primers: 52a, 52b2, temperature: 50C, time: 2:30
PCR for araBAD K808000
template: K808000, primers: 12a, 12bn, temperature: 50C, time: 2:30
PCR to extract Nir2 from P. aeruginosa
PCR for araBAD and SPL with DMSO
Template and primers as previous.
PCR on Nir1 with MgCl2 gradient
Used previous isolation from a gel purification to make PCR for amplification of the strand. Used 5% DMSO as additive. The PCR was done with primer pair 41 and with x7 polymerase at 55C annealing and 5:00 min extension. Also we used 20 sec as denaturing time instead of the normal 10. The magnesium gradient was as follow:
- 0
- 1uL MgCl2 2mM per 50uL reaction
- 5uL MgCl2 2mM per 50uL reaction
- 20uL MgCl2 2mM per 50uL reaction
Gel purification
of the Nir1 fragment extracted from P. aeruginosa. Nanodrop: 8ng/uL
Plasmid miniprep
HAO in pZA21 from USER cloning was purified for restriction analysis. Nanodrop measurement: 180ng/uL
Results
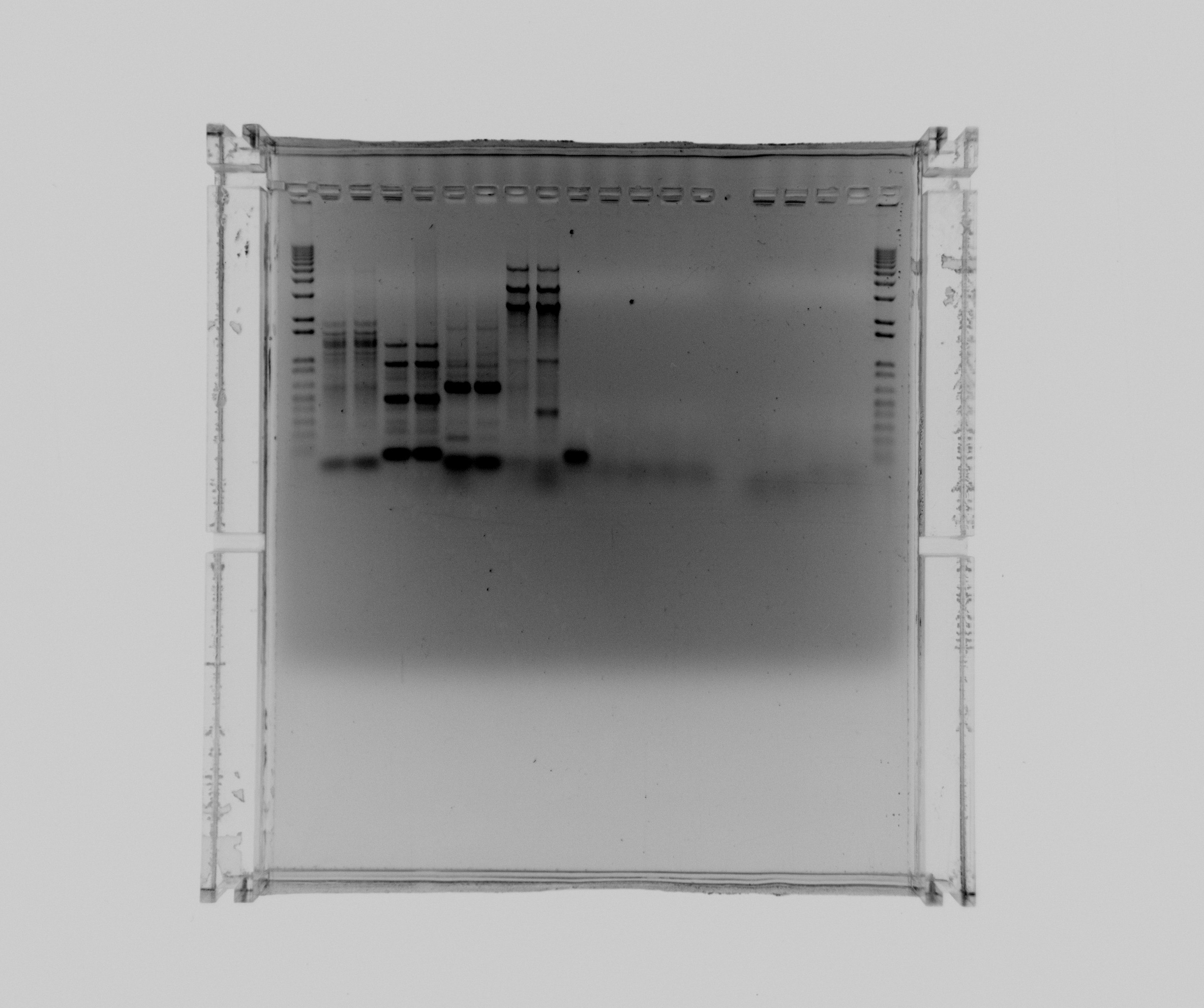
Gel 1
1% agarose gel
- 1 kb ladder
- Nir1 col. Taq
- Nir1 col. Taq
- Nir2 col. Taq
- Nir2 col. Taq
- Nir1 frag. Taq
- Nir1 frag. Taq
- Nir1 col. x7
- Nir1 col. x7
- neg
- Hi spec. buffer
- Hi spec. buffer
- DMSO
- DMSO
- Nir1
- Nir1
- Nir2
- Nir2
- 1 kb ladder
Nir1 was purified.
Gel 2
1% agarose gel
- 1 kb ladder
- NirW
- NirW
- araBAD biobrick K808000
- araBAD biobrick K808000
- SPL in pZA21 containing RFP
- SPL in pZA21 containing RFP
- reference promoter in pZA21 containing RFP
- reference promoter in pZA21 containing RFP
- neg
- neg
- 1 kb ladder
Gel 3
1% agarose gel
- 1 kb ladder
- AMO, 1uL of -20 culture as template
- AMO, 10uL of -20 culture as template
- AMO, 1uL of glycerol stock as template
- AMO, 10uL of glycerol stock as template
- neg from AMO PCR
- 1 - SPL, primers 52a, 52b1, template pZA21 with RFP, 50C, 2:30 min extension
- 2 - SPL, primers 52a, 52b1, template pZA21 with RFP, 50C, 2:30 min extension
- 3 - ref, primers 52a, 52b2, template pZA21 with RFP, 50C, 2:30 min extension
- 4 - ref, primers 52a, 52b2, template pZA21 with RFP, 50C, 2:30 min extension
- 5 - araBAD, primers 12a, 12bn, template K80800, 50C, 2:30 min extension
- 6 - araBAD, primers 12a, 12bn, template K80800, 50C, 2:30 min extension
- 1 kb ladder
AMO was gel purified.
Conclusion
- Nir was obtained from colony PCR and purified.
- AMO was extracted and gel purified.
Navigate to the Previous or the Next Entry
 "
"