Team:Heidelberg/Templates/Del week13 FG
From 2013.igem.org
(Difference between revisions)
(→Restriction digest of fragment from FS_20 to FS_07; 5.2 kb; 23-07-2013 and 24-07-2013) with XmaI) |
(→Restriction digest of fragment from FS_20 to FS_07; 5.2 kb; 23-07-2013 and 24-07-2013 with XmaI) |
||
| Line 433: | Line 433: | ||
==25-07-2013== | ==25-07-2013== | ||
===Restriction digest of fragment from FS_20 to FS_07; 5.2 kb; [[DelF-G#23-07-2013|23-07-2013]] and [[DelF-G#24-07-2013|24-07-2013]] with XmaI=== | ===Restriction digest of fragment from FS_20 to FS_07; 5.2 kb; [[DelF-G#23-07-2013|23-07-2013]] and [[DelF-G#24-07-2013|24-07-2013]] with XmaI=== | ||
| - | [[File:Heidelberg_20130725 FS20toFS07 test restrictionbesch.png|150px|thumb|Restriction digest of Fragment FS_20 to FS_07 (23.07 and 24.07) with XmaI | + | [[File:Heidelberg_20130725 FS20toFS07 test restrictionbesch.png|150px|thumb|Restriction digest of Fragment FS_20 to FS_07 (23.07 and 24.07) with XmaI ; Expected size of digested fragments: 0.9kbp and 4.3kbp; run at 100 V, 0.8 % gel (TAE)]] |
Incubation at 37°C for 45 min | Incubation at 37°C for 45 min | ||
{| class="wikitable" | {| class="wikitable" | ||
Revision as of 17:12, 1 October 2013
Contents[hide] |
22-07-2013
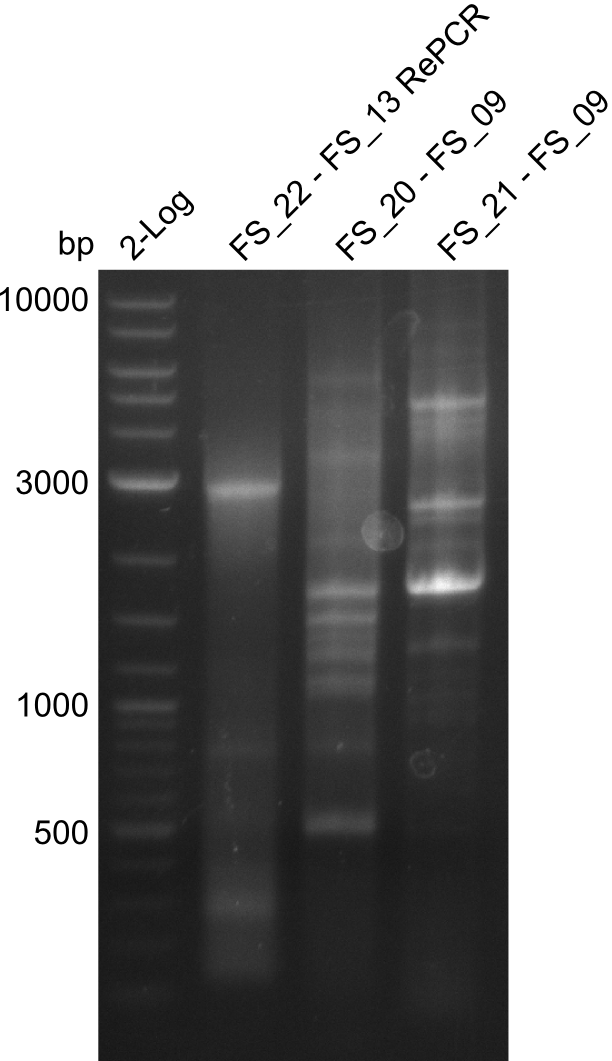
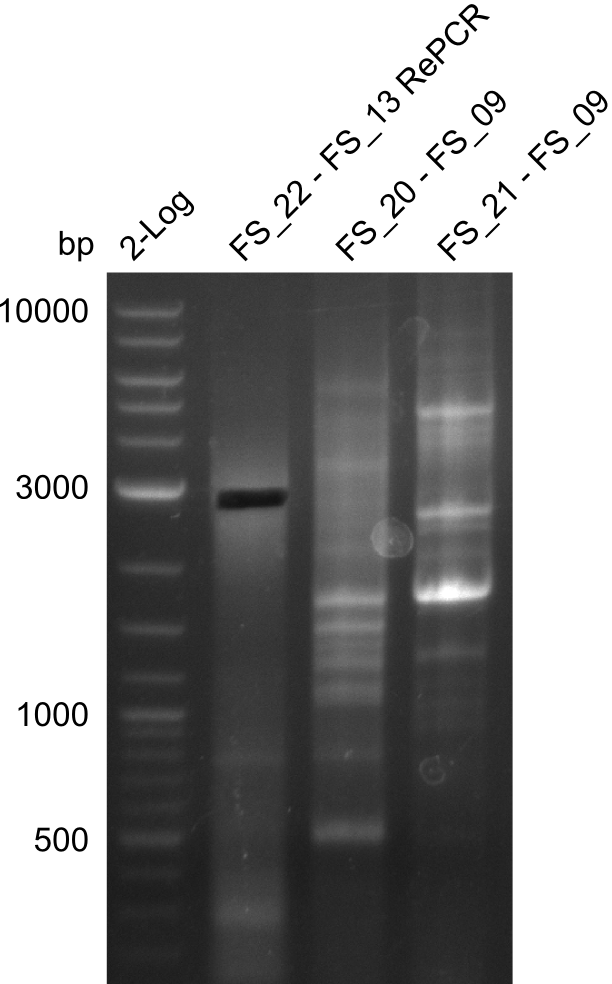
Amplification from FS_20/FS_21 to FS_09; 8.5 kb
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_20/FS_21: (1/10) | 2 |
| FS_09: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 65 ↓ 0.5 | 5 | |
| 72 | 2:20 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 2:20 | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work
- other primers/combinations of primers will be used
- somehow getting annoyed by this part of D. Acidovorans
Amplification from FS_20/FS_21 to FS_11_short; 11.6 kb
4x 20µl (70 touchdown, 65 touchdown)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_20 or FS_21: (1/10) | 2 |
| FS_11_short: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions I
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 70 ↓ 0.5 | 5 | |
| 72 | 3:40 | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 3:40 | |
| 1 | 72 | 13min |
| 1 | 12 | inf |
- Conditions II of Del FG
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 65 ↓ 0.5 | 5 | |
| 72 | 3:40 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 3:40 | |
| 1 | 72 | 13min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work neither with a touchdown PCR starting at an annealing temperature of 65°C nor 70°C
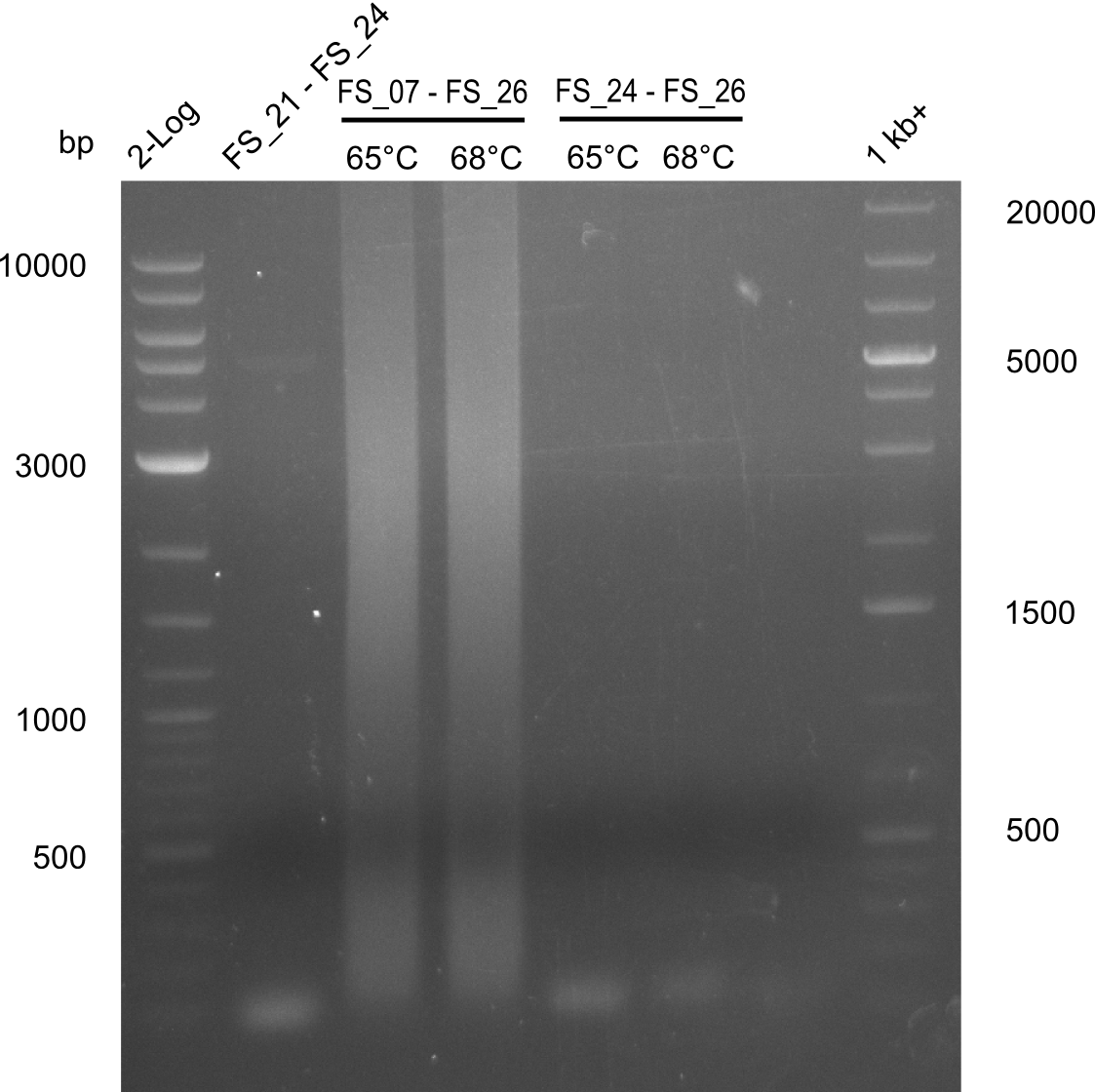
Amplification from FS_20/FS_21 to FS_23; 11.6 kb
4x 20µl (70 touchdown, 65 touchdown)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_20 or FS_21: (1/10) | 2 |
| FS_23: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions I
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 70 ↓ 0.5 | 5 | |
| 72 | 3:40 | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 3:40 | |
| 1 | 72 | 13min |
| 1 | 12 | inf |
- Conditions II
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 65 ↓ 0.5 | 5 | |
| 72 | 3:40 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 3:40 | |
| 1 | 72 | 13min |
| 1 | 12 | inf |
Results:
- Neither the amplification with the primers FS_20 to FS_23 or FS_21 to FS_23 did work. Another primer combination has to be tried.
24-07-2013
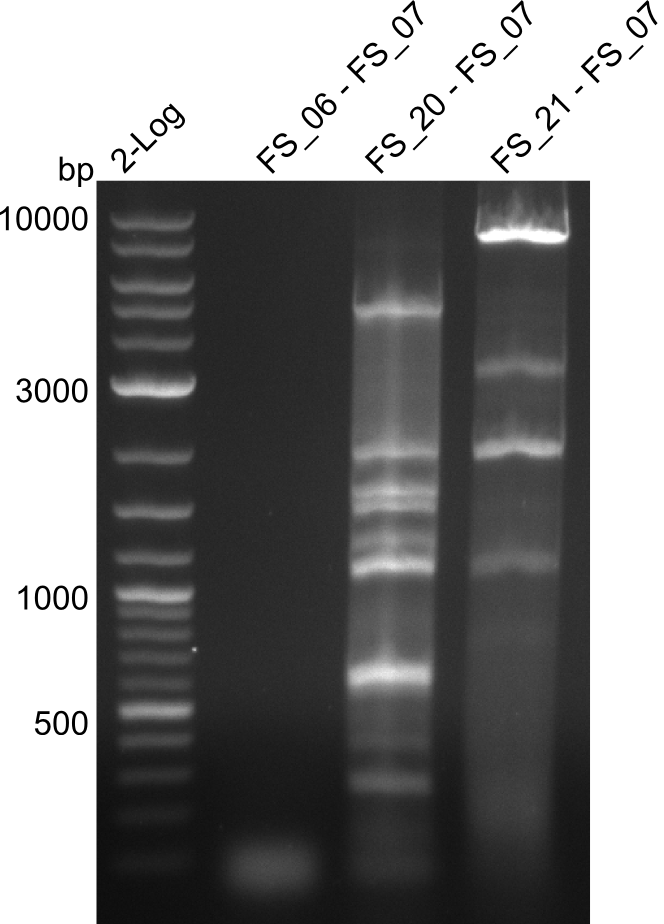
Amplification of DelFG (FS_06 to FS_07; 5.2 kb)
- Reaction I
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_06: (1/10) | 4 |
| FS_07: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
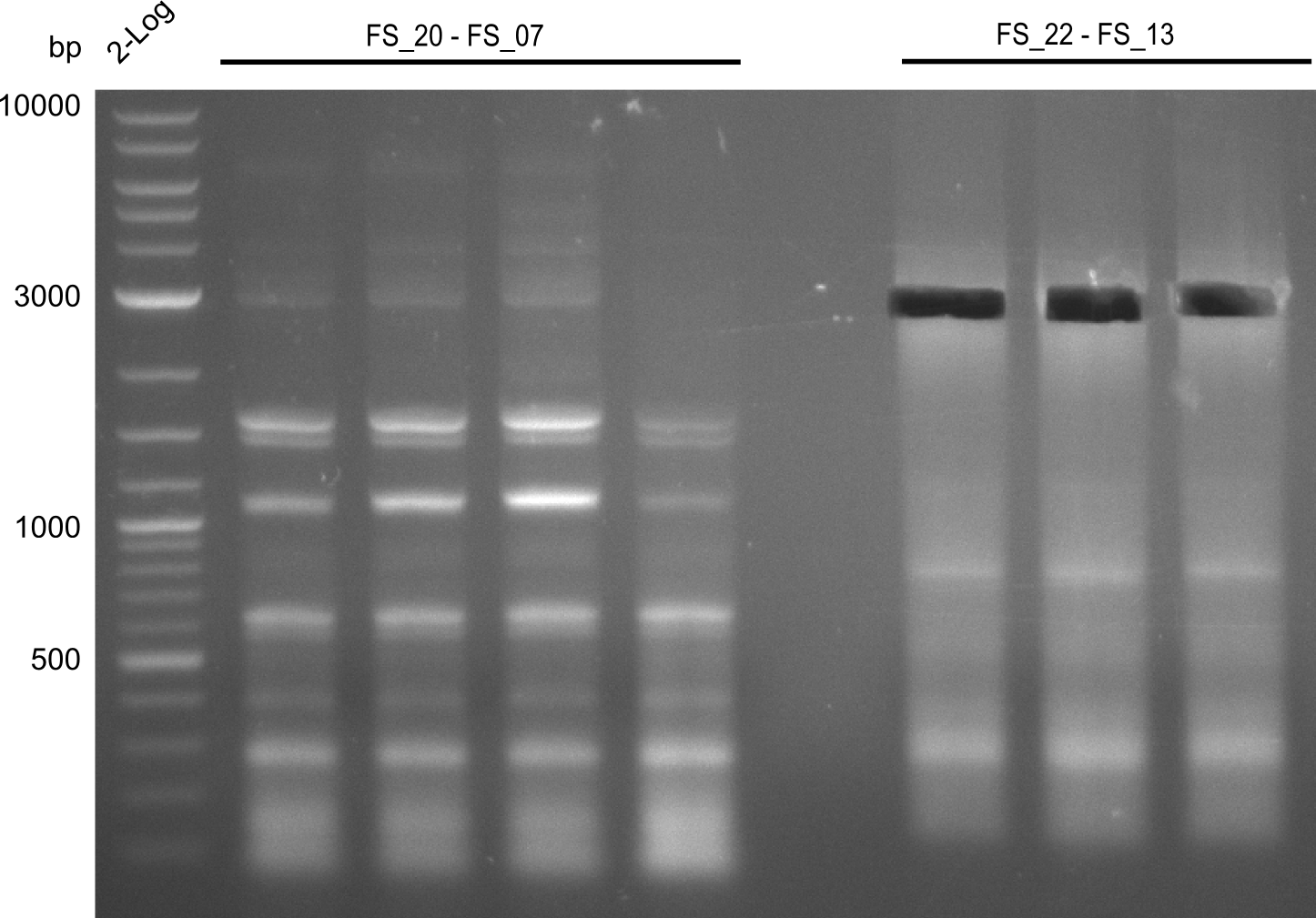
Amplification from FS_20 to FS_07; 5.2 kb
- Reaction II
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_20: (1/10) | 4 |
| FS_07: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
Amplification from FS_21 to FS_07; 5.2kb
- Reaction III
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_21: (1/10) | 4 |
| FS_07: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
- Conditions for reactions I - III
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 2:10 | |
| 18 | 98 | 1 |
| 64 | 5 | |
| 72 | 2:10 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- The amplification of FS_06 to FS_07 did not work. No bands were visible
- The amplification of FS_21 to FS_07 led to several bands, but none of these was the intended product
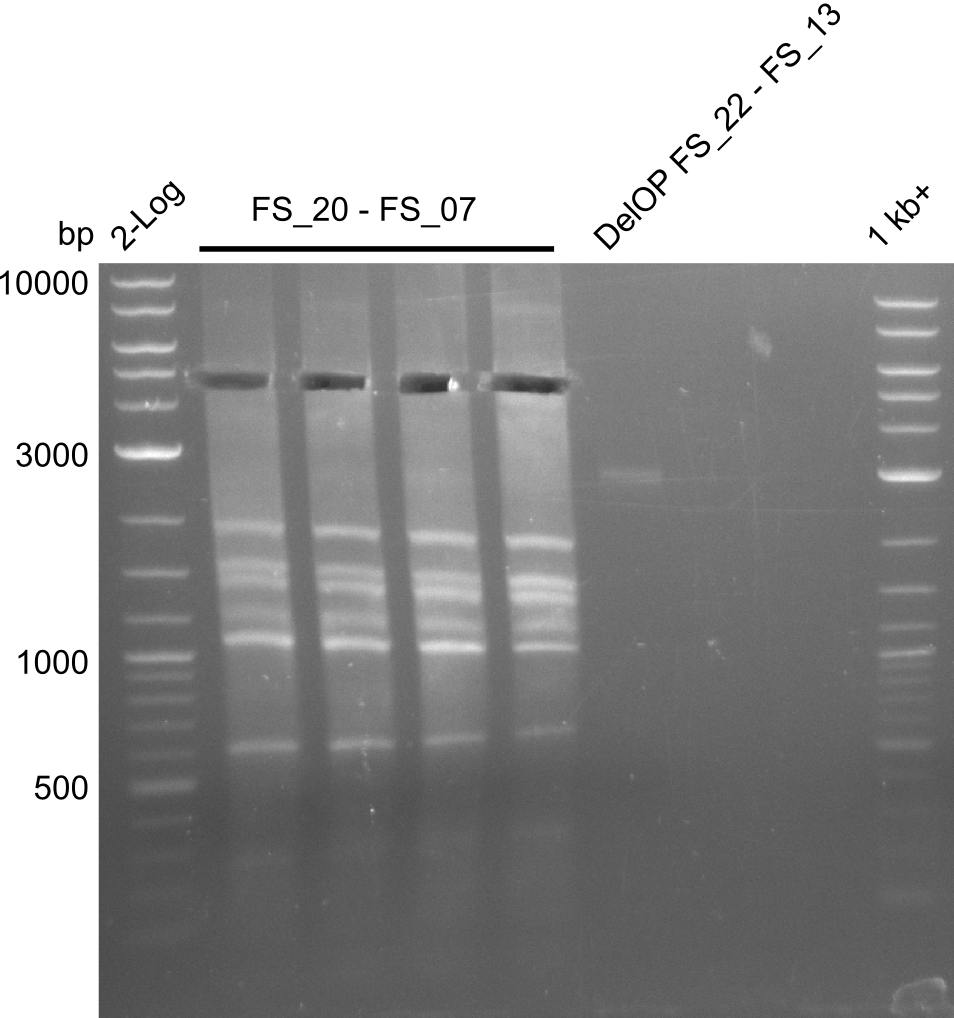
- The amplfication of FS_20 to FS_07 also led to several bands, one band at the right height was observed. Consequently the specificity of the PCR weill be increased by a higher annealing temperature
Amplification from FS_20 to FS_07; 5.2 kb
2x20µl (one with conditions I, other one with conditions II)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_20: (1/10) | 4 |
| FS_07: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
- Conditions I
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 70 ↓ 0.5 | 5 | |
| 72 | 2:10 | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 2:10 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
- Conditions II
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 2:10 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:10 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work, several bands occured, one of these had the size of the intended product but purity of the PCR was not sufficient for Gibson Assembly
- nethertheless bands were cut out and DNA purified using QIAquick Gel Extraction Kit for restriction digest
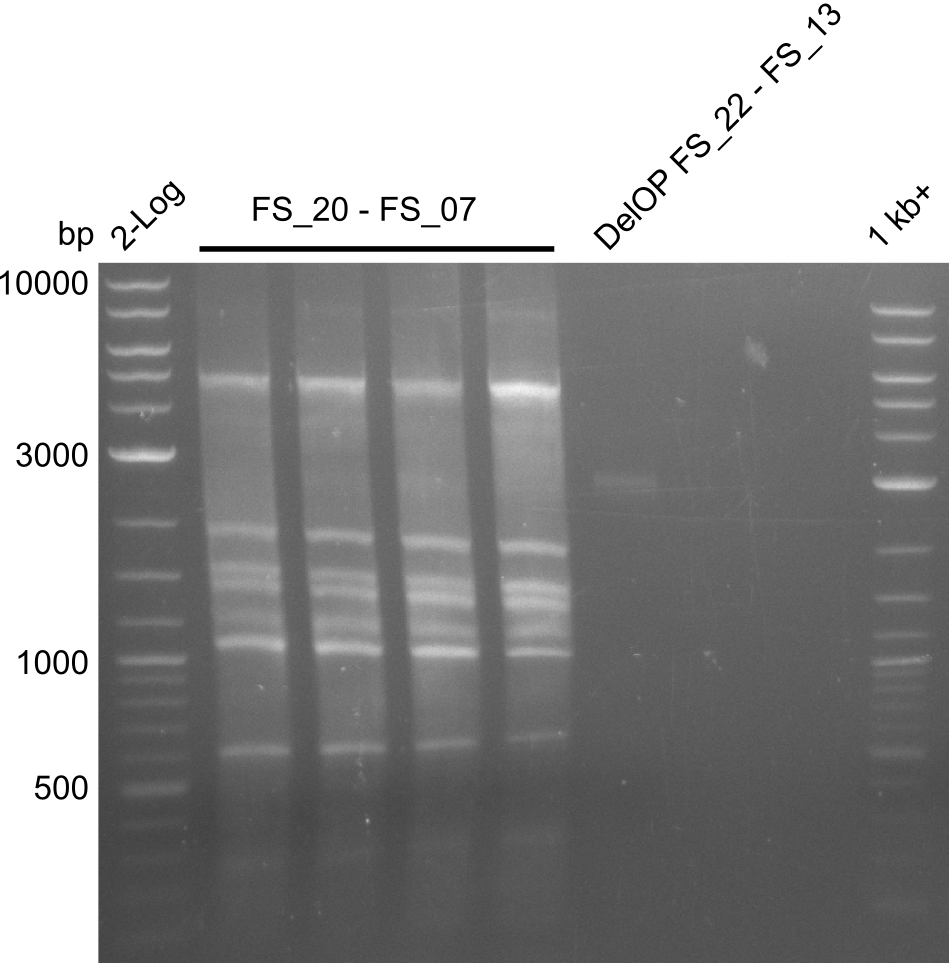
Amplification from FS_20 to FS_07; 5.2 kb
4x20µl
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_20: (1/10) | 4 |
| FS_07: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 70 ↓ 0.5 | 5 | |
| 72 | 2:10 | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 2:10 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work, several bands occured, one of these had the size of the intended product but purity of the PCR was not sufficient for Gibson Assembly
- nethertheless bands were cut out and DNA purified using QIAquick Gel Extraction Kit for restriction digest
25-07-2013
Restriction digest of fragment from FS_20 to FS_07; 5.2 kb; 23-07-2013 and 24-07-2013 with XmaI
Incubation at 37°C for 45 min
| what | µl |
|---|---|
| FS_20 to FS_07 (23-07-2013 and 24-07-2013) | 15 |
| XmaI | 0.8 |
| Buffer CutSmart | 2 |
| dd H2O | 2.2 |
| Expected fragment lengths [bp] | 4307, 879 |
Results:
- restriction digest of DelFG did not work, only very slight bands were visible
- digest will be repeated with higher amount of DNA and enzyme to improve analysis on the gel
26-07-2013
Amplification from FS_20 to FS_07; 5.2 kb
- Reaction I
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_20: (1/10) | 4 |
| FS_07: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 70 | 5 | |
| 72 | 2:10 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
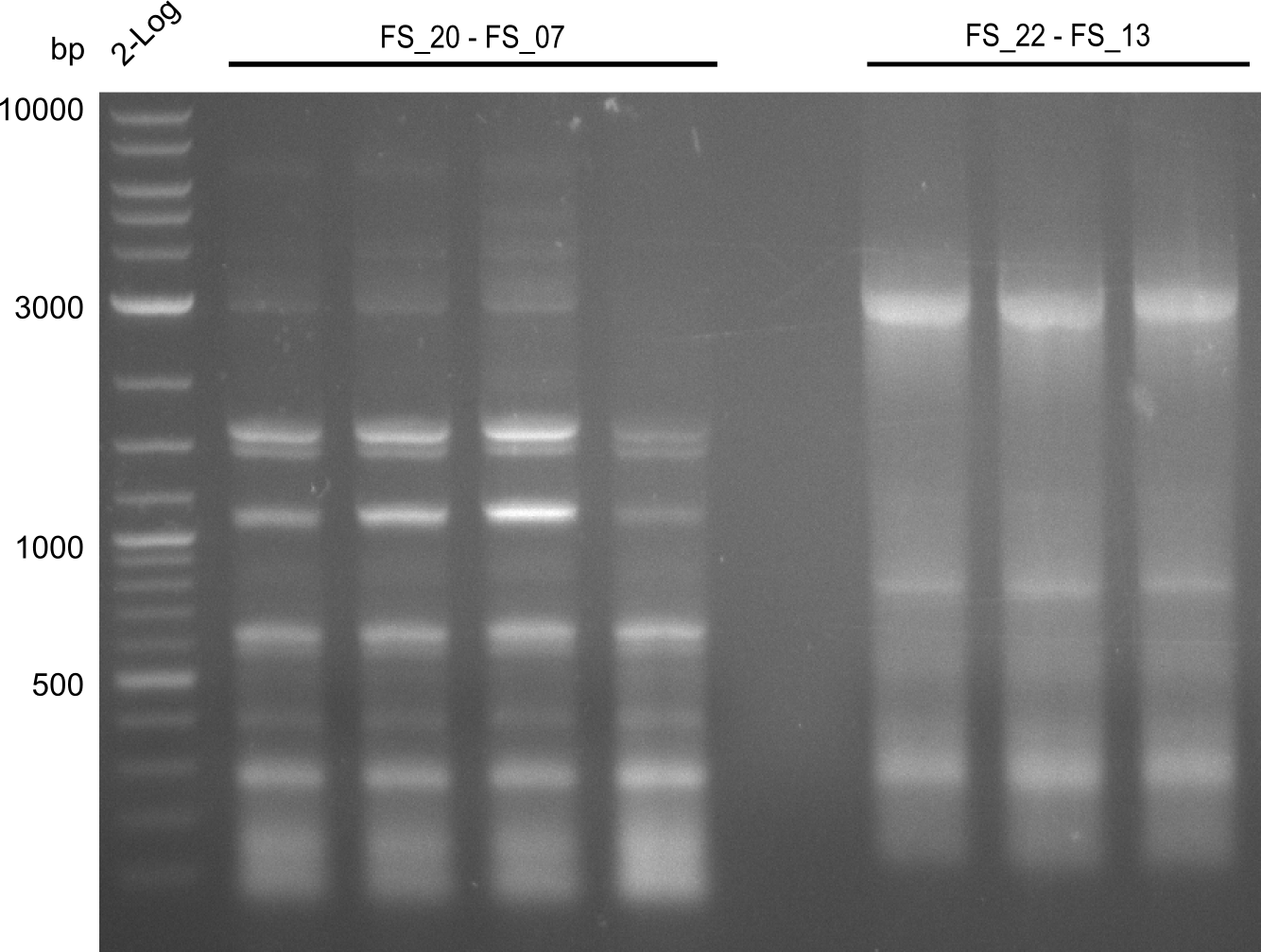
- Amplification of DelFG did not work, several bands occured, one of these had the size of the intended product but purity of the PCR was not sufficient for Gibson Assembly
- Annealing temperature will be increased further, to optimize primer specifity
Restriction digest of fragment from FS_20 to FS_07; 5.2 kb; 23-07-2013 and 23-07-2013) with ClaI
Incubation at 37°C for 45 min
| what | µl |
|---|---|
| FS_20 to FS_07 (23-07-2013 and 23-07-2013) | 25 |
| ClaI | 1 |
| Buffer CutSmart | 3 |
| dd H2O | 1 |
| Expected fragment lengths [bp] | 2743, 1519, 1208 |
Results:
- restriction digest of Del FG did not lead to the expected results
- as no DNA was visible in the restriction digest, experiment will be repeated with a higher amount of DNA
Amplification from FS_06 to FS_07; 5.2 kb
4 x 20µL
- Reaction I
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_20: (1/10) | 4 |
| FS_07: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 72 ↓ 0.5 | 5 | |
| 72 | 2:10 | |
| 18 | 98 | 1 |
| 70 | 5 | |
| 72 | 2:10 | |
| 1 | 72 | 10min |
| 1 | 12 | inf |
Results:
- Amplification of DelFG did not work, several bands occured, one of these had the size of the intended product but purity of the PCR was not sufficient for Gibson Assembly
- nethertheless bands were cut out and DNA purified using QIAquick Gel Extraction Kit for restriction digest
- Does anyone know, why we are constantly repeating this totally deficient PCR?
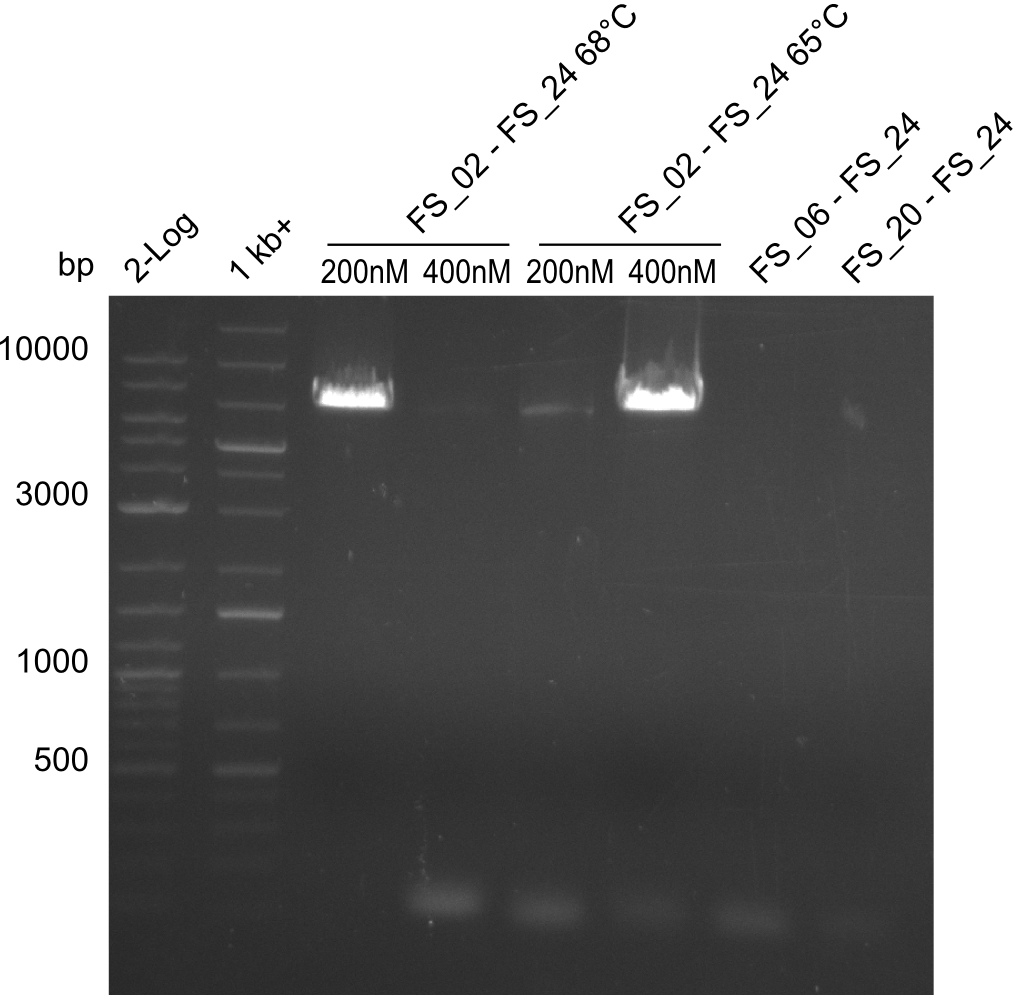
Amplification from FS_6/FS_20/FS_21 to FS_24 (PRIMER FS_24 MIXED UP!)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_06/FS_20/FS_21: (1/10) | 2 |
| FS_24: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 1:20 | |
| 1 | 72 | 7 min |
| 1 | 10 | inf |
Results:
- no PCR product occured since the wrong primers were used
28-07-2013
Amplification from FS_21 to FS_24; (PRIMER 24 was MIXED UP!)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_21: (1/10) | 2 |
| FS_24: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 2:30 | |
| 1 | 72 | 8 min |
| 1 | 12 | inf |
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_21: (1/10) | 2 |
| FS_24: (1/10) | 2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 4 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 60 | 5 | |
| 72 | 2:30 | |
| 1 | 72 | 8 min |
| 1 | 12 | inf |
Results:
- no PCR product occured since the wrong primers were used
 "
"