Team:Heidelberg/Template/Del week17 Gibson
From 2013.igem.org
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | |||
==19-08-2013== | ==19-08-2013== | ||
===Colony PCRs for testing ligation of pSB4K5 and DelL=== | ===Colony PCRs for testing ligation of pSB4K5 and DelL=== | ||
| Line 96: | Line 95: | ||
<gallery> | <gallery> | ||
| - | File:Heidelberg_Sequencing Result pFSN FS 13s colony D8w.clustal | Alignment for sequencing of pFSN with primer FS_13s_rv against the expected construct to validate sequence of DelOP | + | File:Heidelberg_Sequencing Result pFSN FS 13s colony D8w.clustal.txt | Alignment for sequencing of pFSN with primer FS_13s_rv against the expected construct to validate sequence of DelOP |
| - | File:Heidelberg_Sequencing Result pFSN FS 57 colony D8w.clustal | Alignement for sequencing of pFSN with primer FS_57_rv against the expected construct to validate the sequence for the overlap of DelOP to DelG | + | File:Heidelberg_Sequencing Result pFSN FS 57 colony D8w.clustal.txt | Alignement for sequencing of pFSN with primer FS_57_rv against the expected construct to validate the sequence for the overlap of DelOP to DelG |
| - | File:Heidelberg_Sequencing Result pFSN FS 58 colony D8w.clustal | Alignement for sequencing of pFSN with primer FS_58_fw against the expected construct to validate the sequence for the overlap of DelOP to DelL | + | File:Heidelberg_Sequencing Result pFSN FS 58 colony D8w.clustal.txt | Alignement for sequencing of pFSN with primer FS_58_fw against the expected construct to validate the sequence for the overlap of DelOP to DelL |
| - | File:Heidelberg_Sequencing Result pFSN FS 63 colony D8w.clustal | Alignement for sequencing of pFSN with primer FS_63_rv against the expected construct to validate the sequence for the overlap of DelL to mRFP | + | File:Heidelberg_Sequencing Result pFSN FS 63 colony D8w.clustal.txt | Alignement for sequencing of pFSN with primer FS_63_rv against the expected construct to validate the sequence for the overlap of DelL to mRFP |
| - | File:Heidelberg_Sequencing Result pFSN VF2 colony D8w.clustal | Alignement for sequencing of pFSN with primer VF2 against the expected construct to validate the sequence for the overlap of pSB4K5 to DelAF | + | File:Heidelberg_Sequencing Result pFSN VF2 colony D8w.clustal.txt | Alignement for sequencing of pFSN with primer VF2 against the expected construct to validate the sequence for the overlap of pSB4K5 to DelAF |
</gallery> | </gallery> | ||
Revision as of 14:05, 3 October 2013
Contents |
19-08-2013
Colony PCRs for testing ligation of pSB4K5 and DelL
- Reaction mixture
| Reagent | E13w | E14w | E15w | F25w | F26w | F27w |
|---|---|---|---|---|---|---|
| VR-Primer: (1/10) | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl |
| FS_14: (1/10) | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl | 2 µl |
| Colonies | Colony E13w liquid culture | Colony E14w liquid culture | Colony E15w liquid culture | Colony F25w liquid culture | Colony F26w liquid culture | Colony F27w liquid culture |
| DreamTaq | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| dd H2O | 5 µl | 5 µl | 5 µl | 5 µl | 5 µl | 5 µl |
Result:
- Colony F26w was screened positive, as it shows the expected band at 1.4kbp, the others were screened negative
- 8 ml of colony D8w and colony D26w (pFSN-1) were midi-preped according to protocol 'Preparation of large plasmids'.
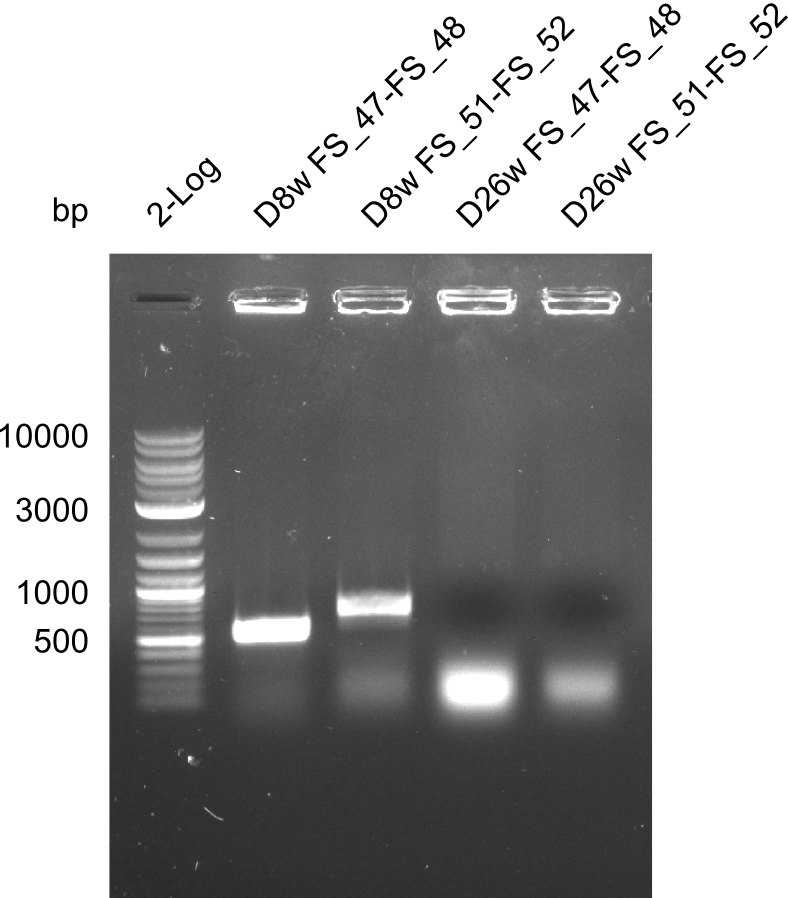
Colony PCRs for testing ligation of pSB4K5 and DelAF, as well as DelAF and DelFG
- Reaction mixture
| Reagent | D8w | D8w | F26w | F26w |
|---|---|---|---|---|
| Primer_fw (1/10) | 2 µl FS_47 | 2 µl FS_51 | 2 µl FS_47 | 2 µl FS_51 |
| Primer_rv (1/10) | 2 µl FS_48 | 2 µl FS_52 | 2 µl FS_48 | 2 µl FS_52 |
| Colonies | Colony D8w liquid culture | Colony D8w liquid culture | Colony F26w liquid culture | Colony F26w liquid culture |
| DreamTaq | 10 µl | 10 µl | 10 µl | 10 µl |
| dd H2O | 5 µl | 5 µl | 5 µl | 5 µl |
| Expected band | 547 bp | 724 bp | 547 bp | 724 bp |
Results:
- The screening was positive for clone D8w with both primer combinations.
- Thus this clone contains the backbone pSB4K5, the genes DelA, DelB, DelC, DelD, DelE, DelF, DelL and at least a part of the gene DelG.
- For colony F26w the screenings were negativ.
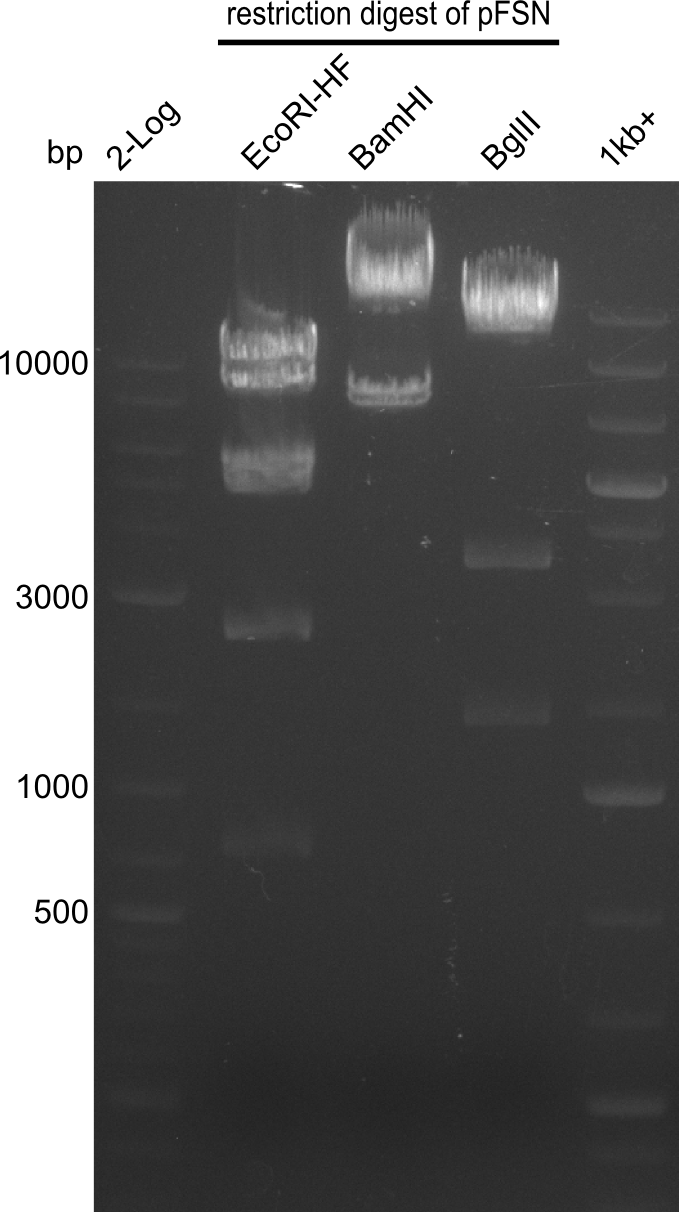
Restriction digest of pFSN (31.435 kbp)
Incubation at 37°C for about 6 hours
| Reagent | µl | ||
|---|---|---|---|
| pFSN | 1 | ||
| EcoRI | BamH1 | BglI | 2 |
| CutSmart Buffer | 0.5 | ||
| dd H2O | 1.5 | ||
| Expected fragment lengths [bp] EcoRI | 9798, 7856, 5276, 4624, 2529, 1212 | ||
| Expected fragment lengths [bp] BamHI | 23860, 7435 | ||
| Expected fragment lengths [bp] BglII | 14223, 11919, 3291, 1892 | ||
Results:
- All digests displayed the expected bands and thus were positive.
- Therefore the pFSN plasmid is very likely our desired construct.
20-08-2013
Sequencing
- The construct pFSN was isolated from overnight culture using the 'Preparation of large plasmids' protocol.
- Single read sequencing was carried out by GATC Biotech
- obtained .abi files were analyzed with UniPro Ugene, part of sequence showing convenient chromatogram was chosen for alignment using ClustalOmega
Sequence-Files from GATC Biotech
Alignments
Results:
- Sequencing of the final construct pFSN for all internal Gibson sites validated the intended sequence
- Sequence of mRFP shows mutations within the primer site, therefore it can be concluded that mRFP is not functional
- Colonies of D8w will be further analyzed by FACs to investigate whether mRFP is functional or not
- Additionaly, SDS-page of clone D8w will be conducted to check for expression of the inserted Del genes
- Concluding the results of colony PCRs, restriction digests and sequencing it can be concluded, that we successfully amplified 28 kbp of genomic DNA from our donator organism Delftia acidovorans SPH-1 distributed from the DSMZ, ligated the thereby obtained 6 fragments including the standard biobrick backbone pSB4K5 sucessfully in the intended order and consequently transformed a plasmid of 32 kbp in total into E. Coli using electroporation
21-08-2013
- Another 8 ml of colony D8w (pFSN-1) were midi-preped according to protocol 'Preparation of large plasmids'.
 "
"