Team:TU-Munich/Modeling/Kill Switch
From 2013.igem.org
ChristopherW (Talk | contribs) (→siRNA Model) |
ChristopherW (Talk | contribs) (→siRNA Model) |
||
| Line 18: | Line 18: | ||
====siRNA Model==== | ====siRNA Model==== | ||
| - | We determined the governing equations of this model to be the following: | + | We determined the governing equations of this model to be the following:[[File:TUM13_siRNA_formula.png|center]] |
| - | [[File:TUM13_siRNA_formula.png|center]] | + | |
with initial conditions V(0) = 1 and R(0) = 0, where at the time t=0 the trigger is activated. | with initial conditions V(0) = 1 and R(0) = 0, where at the time t=0 the trigger is activated. | ||
| Line 45: | Line 44: | ||
| - | + | So there is only one stable point in this range, namely: | |
[[File:TUM13_siRNA_stable_realistic_point.png|center]] | [[File:TUM13_siRNA_stable_realistic_point.png|center]] | ||
| - | [[File:TUM13_siRNA_expandby.png | + | By expanding the fraction by [[File:TUM13_siRNA_expandby.png]] we can rewrite V* as |
| - | + | ||
[[File:TUM13_siRNA_V_rewritten.png|center]] | [[File:TUM13_siRNA_V_rewritten.png|center]] | ||
| - | |||
| - | [[File:TUM13_siRNA_Hessian.png|center]] | + | So [[File:TUM13_siRNA_V_in01.png]]. |
| + | |||
| + | |||
| + | Now look at the eigenvalues of the Hessian matrix to analyze the stability [[File:TUM13_siRNA_Hessian.png|center]] | ||
| - | |||
| + | Defining [[File:TUM13_siRNA_EigVals_b.png]], the eigenvalues are given by | ||
[[File:TUM13_siRNA_EigVals.png|center]] | [[File:TUM13_siRNA_EigVals.png|center]] | ||
| - | [[File:TUM13_siRNA_END.png | + | So [[File:TUM13_siRNA_END.png]], which means that this is a stable attractor. |
====Nuclease Modell==== | ====Nuclease Modell==== | ||
Revision as of 15:04, 3 October 2013
Kill Switch Modeling
Purpose
The idea of our kill switch is to kill off our moss, as soon as it leaves the filter system. For this purpose two methods were proposed:
- siRNA method: When some trigger is activated, siRNA is expressed inhibiting the expression of a vital gene
- nuclease method: When some trigger is activated, a nuclease is released destroying the DNA of the cell
To decide between these two methods we modelled the vitality V of the cell (a number between 0 and 1, so a perfectly functional cell has V=1, a dead cell V=0) and depending on the tested method the concentration of siRNA R and nuclease N as appropriate. Both concentrations are normalized to the unit interval [0,1].
siRNA Model
We determined the governing equations of this model to be the following:with initial conditions V(0) = 1 and R(0) = 0, where at the time t=0 the trigger is activated.
The stable points V* and R* of this system have to satisfy Defining
If α = 1, the unique stable point is ![]() .
.
These are both negative, so this is a stable point.
Now only one of these is in the sensible range, because
So there is only one stable point in this range, namely:
By expanding the fraction by ![]() we can rewrite V* as
we can rewrite V* as
Defining ![]() , the eigenvalues are given by
, the eigenvalues are given by
So ![]() , which means that this is a stable attractor.
, which means that this is a stable attractor.
Nuclease Modell
Conclusion
For a functional kill-switch it is necessary, that the cells are actually killed completely and not just live on with reduced vitality. So based on our modelling results the siRNA approach is not satisfactory, while the nuclease satisfies the requirement. As a result the team pursued the nuclease approach leading to our final kill-switch.
 "
"



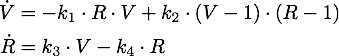
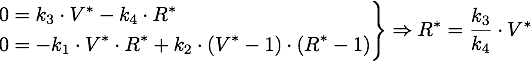
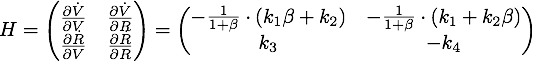
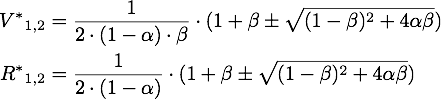
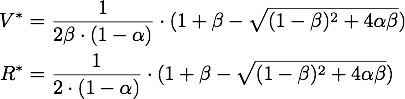
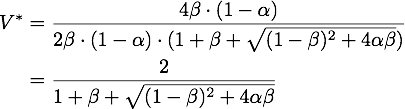
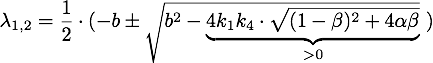
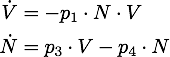

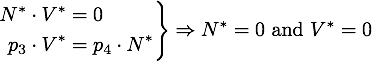
AutoAnnotator:
Follow us:
Address:
iGEM Team TU-Munich
Emil-Erlenmeyer-Forum 5
85354 Freising, Germany
Email: igem@wzw.tum.de
Phone: +49 8161 71-4351