Team:INSA Toulouse/contenu/lab practice/results/polT7
From 2013.igem.org
(Difference between revisions)
| Line 157: | Line 157: | ||
<h2 class="title2">Conception</h2> | <h2 class="title2">Conception</h2> | ||
| - | The following constructions were designed:<br> | + | The following constructions were designed:<br><br> |
| - | <img src="https://static.igem.org/mediawiki/2013/0/07/Result_polT7_1.jpg" class="imgcenter" /><br> | + | <img src="https://static.igem.org/mediawiki/2013/0/07/Result_polT7_1.jpg" class="imgcenter" /><br><br> |
Coding sequence of T7 polymerase was extracted from the genome of E.Coli BL21-DE3. Cloning are necessary to add a promoter, a rbs and a terminator. The expected functioning is: | Coding sequence of T7 polymerase was extracted from the genome of E.Coli BL21-DE3. Cloning are necessary to add a promoter, a rbs and a terminator. The expected functioning is: | ||
- Production of RFP if T7 polymerase is present (red colonies) | - Production of RFP if T7 polymerase is present (red colonies) | ||
| Line 167: | Line 167: | ||
Nevertheless, a new biobrick pT7-RFP was created! | Nevertheless, a new biobrick pT7-RFP was created! | ||
| - | Following is the electrophoresis gel showing that we obtained a fragment of expected length (829 bp) by restriction EcoRI/PstI:<br> | + | Following is the electrophoresis gel showing that we obtained a fragment of expected length (829 bp) by restriction EcoRI/PstI:<br><br> |
| - | <img src="https://static.igem.org/mediawiki/2013/f/f9/Result_polT7_2.jpg" class="imgcenter" /><br> | + | <img src="https://static.igem.org/mediawiki/2013/f/f9/Result_polT7_2.jpg" class="imgcenter" /><br><br> |
Further verifications were done with phenotypic test. Plasmid was transformed into competent BL21-DE3. BL21-DE3 is the strain containing T7 polymerase under of Lac promoter (inductible with IPTG). | Further verifications were done with phenotypic test. Plasmid was transformed into competent BL21-DE3. BL21-DE3 is the strain containing T7 polymerase under of Lac promoter (inductible with IPTG). | ||
| - | White colonies was obtained into petri dish and red colonies into petri dish with IPTG <br> | + | White colonies was obtained into petri dish and red colonies into petri dish with IPTG <br><br> |
<img src="https://static.igem.org/mediawiki/2013/4/43/Result_polT7_3.jpg" class="imgcenter" /><br> | <img src="https://static.igem.org/mediawiki/2013/4/43/Result_polT7_3.jpg" class="imgcenter" /><br> | ||
Revision as of 20:12, 3 October 2013
Results - T7 Polymerase Characterization
Objective
Characterize the activity of T7 polymerase with an RFP reporter under a T7 promoter. Reminder: T7 polymerase is necessary in our system to activate the two T7 promoters in the AND1 and AND2 gate.Conception
The following constructions were designed: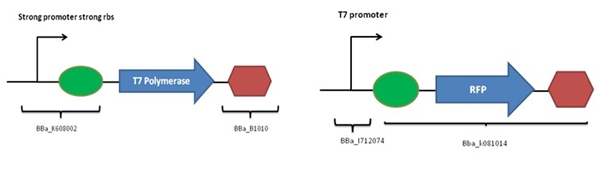
Coding sequence of T7 polymerase was extracted from the genome of E.Coli BL21-DE3. Cloning are necessary to add a promoter, a rbs and a terminator. The expected functioning is: - Production of RFP if T7 polymerase is present (red colonies) - Absence of RFP if T7 polymerase is absent (white colonies)
Result
Cloning to add a promoter and a rbs to T7 polymerase was a success but Cloning for adding a terminator to T7 polymerase was never achieved. Nevertheless, a new biobrick pT7-RFP was created! Following is the electrophoresis gel showing that we obtained a fragment of expected length (829 bp) by restriction EcoRI/PstI:
Further verifications were done with phenotypic test. Plasmid was transformed into competent BL21-DE3. BL21-DE3 is the strain containing T7 polymerase under of Lac promoter (inductible with IPTG). White colonies was obtained into petri dish and red colonies into petri dish with IPTG


Dicussion
The biobrick behave as expected. It was submitted to the registry (lien BBa_K1132045). Further characterization can be done concerning the activity of T7 polymerase, looking to the production of RFP during time.
 "
"

