Team:UNIK Copenhagen/Notebook
From 2013.igem.org
SigneKarlsen (Talk | contribs) |
SigneKarlsen (Talk | contribs) |
||
| (113 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:UNIK_Copenhagen/template}} | {{:Team:UNIK_Copenhagen/template}} | ||
<html> | <html> | ||
| - | |||
<link href='http://fonts.googleapis.com/css?family=The+Girl+Next+Door' rel='stylesheet' type='text/css'> | <link href='http://fonts.googleapis.com/css?family=The+Girl+Next+Door' rel='stylesheet' type='text/css'> | ||
| - | |||
<style type="text/css"> | <style type="text/css"> | ||
| + | #wrapper{ | ||
| + | height: 1290px; | ||
| + | } | ||
#indent_content{ | #indent_content{ | ||
margin: 30px; | margin: 30px; | ||
| Line 35: | Line 36: | ||
.content img{ | .content img{ | ||
float: left; | float: left; | ||
| - | margin: 10px;} | + | margin: 10px; |
| + | } | ||
.left_page, .right_page{ | .left_page, .right_page{ | ||
float: left; | float: left; | ||
| Line 57: | Line 59: | ||
margin: 0px; | margin: 0px; | ||
width: 46px; | width: 46px; | ||
| - | height: | + | height: 42px; |
} | } | ||
#tabs li a{ | #tabs li a{ | ||
| Line 73: | Line 75: | ||
border: 1px solid gray; | border: 1px solid gray; | ||
border-left: none; | border-left: none; | ||
| - | padding: 9px 14px | + | padding: 9px 14px 28px 14px; |
} | } | ||
#tabs li a:hover{ | #tabs li a:hover{ | ||
| Line 86: | Line 88: | ||
border-left: none; | border-left: none; | ||
} | } | ||
| - | #content2, #content3, #content4, #content5, #content6, #content7, #content8, #content9, #content10, #content11, #content12{ | + | #content2, #content3, #content4, #content5, #content6, #content7, #content8, #content9, #content10, #content11, #content12, #content13{ |
display: none; | display: none; | ||
padding: 10px; | padding: 10px; | ||
| Line 92: | Line 94: | ||
#content4 p, #content7 p{ | #content4 p, #content7 p{ | ||
font-size: 12px; | font-size: 12px; | ||
| + | } | ||
| + | #under{ | ||
| + | width: 880px; | ||
| + | height: 400px; | ||
| + | float:left; | ||
| + | margin-left: -50px; | ||
} | } | ||
#under_page{ | #under_page{ | ||
| - | float:left; | + | margin: 10px; |
| + | width: 200px; | ||
| + | float: left; | ||
| + | } | ||
| + | #under_page2{ | ||
| + | margin: 10px; | ||
| + | width: 180px; | ||
| + | float: left; | ||
| + | } | ||
| + | #confocals{ | ||
| + | height: 170px; | ||
| + | width: 340px; | ||
| + | float: left; | ||
| + | } | ||
| + | #confocals img{ | ||
| + | margin: 5px; | ||
| + | } | ||
| + | #confocal{ | ||
| + | overflow: hidden; | ||
| + | height: 160px; | ||
| + | width: 160px; | ||
| + | float: left; | ||
| + | } | ||
| + | #confocal img:hover{ | ||
| + | margin-top: -165px; | ||
| + | } | ||
| + | #middle{ | ||
| + | margin: 80px 50px 50px 55px; | ||
} | } | ||
</style> | </style> | ||
| Line 100: | Line 135: | ||
<script type="text/javascript"> | <script type="text/javascript"> | ||
function ThisTab(new_tab, new_content) { | function ThisTab(new_tab, new_content) { | ||
| - | for(var i=1;i< | + | for(var i=1;i<14;i++){document.getElementById('content'+i).style.display = 'none';} |
document.getElementById(new_content).style.display = 'block'; | document.getElementById(new_content).style.display = 'block'; | ||
| - | for(var i=1;i< | + | for(var i=1;i<14;i++){document.getElementById('tab'+i).className = '';} |
document.getElementById(new_tab).className = 'active'; | document.getElementById(new_tab).className = 'active'; | ||
} | } | ||
| Line 111: | Line 146: | ||
// Popup window code | // Popup window code | ||
function newPopup(url) { | function newPopup(url) { | ||
| - | popupWindow = window.open( | + | popupWindow = window.open( url,'popUpWindow','height=700,width=800,left=30,top=20,resizable=yes,scrollbars=yes,toolbar=yes,menubar=no,location=no,directories=no,status=yes') |
| - | + | ||
} | } | ||
</script> | </script> | ||
| Line 119: | Line 153: | ||
<h1> Notebook </h1> | <h1> Notebook </h1> | ||
<h4>Get our Nootebook in a <a href="JavaScript:newPopup('https://2013.igem.org/wiki/index.php?title=Team:UNIK_Copenhagen/Notebook/Printable&printable=yes');"><u>printable version here</a></u>.</h4> | <h4>Get our Nootebook in a <a href="JavaScript:newPopup('https://2013.igem.org/wiki/index.php?title=Team:UNIK_Copenhagen/Notebook/Printable&printable=yes');"><u>printable version here</a></u>.</h4> | ||
| - | <p>If nothing happens when you click, check if you have disabled popup-windows.</p> | + | <p>If nothing happens when you click, check if you have disabled popup-windows.<br> For more detailed descriptions of the methods used, please see our <a href="https://static.igem.org/mediawiki/2013/9/99/UNIK_Copenhagen_Protocols.pdf" target="_blank">protocols</a>.</p> |
| - | + | ||
</div> | </div> | ||
| Line 335: | Line 368: | ||
<p> | <p> | ||
<u>Assembly of constructs</u> | <u>Assembly of constructs</u> | ||
| - | Plasmids from last week eFbFP/eGFP-pBBR1MCS-2 were digested alongside the newly assembled MamC-pJET in order to assess whether the restriction sites were present or not ( | + | Plasmids from last week eFbFP/eGFP-pBBR1MCS-2 were digested alongside the newly assembled MamC-pJET in order to assess whether the restriction sites were present or not (see gel no. 1). |
All lanes with eGFP or MamC were positive (see gel above) and the bands were purified from the gel. From this purification very poor concentration was obtained. From the lanes with eFbFP, no. 6 stands out from the negative control. Therefore we decided we continue working with this plasmid. A re-ligation of eFbFP-pBBR1MCS-2 was performed. | All lanes with eGFP or MamC were positive (see gel above) and the bands were purified from the gel. From this purification very poor concentration was obtained. From the lanes with eFbFP, no. 6 stands out from the negative control. Therefore we decided we continue working with this plasmid. A re-ligation of eFbFP-pBBR1MCS-2 was performed. | ||
<br> | <br> | ||
| - | Cloning of MamC into linearized eGFP-pBBR1MCS-2 and subsequently transforming the plasmid was also done. Colony PCR was carried out on MamC-eGFP-pBBR1MCS-2 and eFbFP-pBBR1MCS-2. We decided to use the Gateway (GTW) system to express our constructs ( | + | Cloning of MamC into linearized eGFP-pBBR1MCS-2 and subsequently transforming the plasmid was also done. Colony PCR was carried out on MamC-eGFP-pBBR1MCS-2 and eFbFP-pBBR1MCS-2. We decided to use the Gateway (GTW) system to express our constructs (see gel no. 2). |
To save time the colony PCR was performed using gateway primers (MamC F and eGFP R, eFbFP F and R). The PCR product of MamC-eGFP-pBBR1MCS-2 colonies 1, 3, 4, 5, 6 and eFbFP-pBBR1MCS-2 3, 7, 10, 13, 15 were PCR purified. | To save time the colony PCR was performed using gateway primers (MamC F and eGFP R, eFbFP F and R). The PCR product of MamC-eGFP-pBBR1MCS-2 colonies 1, 3, 4, 5, 6 and eFbFP-pBBR1MCS-2 3, 7, 10, 13, 15 were PCR purified. | ||
| Line 345: | Line 378: | ||
</p> | </p> | ||
| - | <div> | + | <br><br><br> |
| + | <div id="under"> | ||
<div id="under_page"> | <div id="under_page"> | ||
| - | <img src="https://static.igem.org/mediawiki/2013/9/94/UNIK_Copenhagen_Notebook_ref_16.06.jpg" width=" | + | <a href="https://static.igem.org/mediawiki/2013/9/94/UNIK_Copenhagen_Notebook_ref_16.06.jpg" target="_blank"><img src="https://static.igem.org/mediawiki/2013/9/94/UNIK_Copenhagen_Notebook_ref_16.06.jpg" width="200"></a><br> |
<p> Gel no. 1: Digestion of eGFP-pBBR1MCS-2, eFbFP-pBBR1MCS-2 and MamC-pJET </p> | <p> Gel no. 1: Digestion of eGFP-pBBR1MCS-2, eFbFP-pBBR1MCS-2 and MamC-pJET </p> | ||
</div> | </div> | ||
<div id="under_page"> | <div id="under_page"> | ||
| - | <img src="https://static.igem.org/mediawiki/2013/9/9d/UNIK_Copenhagen_Notebook_ref_16.21.jpg" width=" | + | <a href="https://static.igem.org/mediawiki/2013/9/9d/UNIK_Copenhagen_Notebook_ref_16.21.jpg" target="_blank"> <img src="https://static.igem.org/mediawiki/2013/9/9d/UNIK_Copenhagen_Notebook_ref_16.21.jpg" width="200"></a><br> |
<p> Gel no. 2: Colony PCR of MamC-eGFP-pBBR1MCS-2 and eFbFP-pBBR1MCS-2 using Gateway primers </p> | <p> Gel no. 2: Colony PCR of MamC-eGFP-pBBR1MCS-2 and eFbFP-pBBR1MCS-2 using Gateway primers </p> | ||
</div> | </div> | ||
<div id="under_page"> | <div id="under_page"> | ||
| - | <img src="https://static.igem.org/mediawiki/2013/2/2b/UNIK_Copenhagen_Notebook_ref_16.35.jpg" width=" | + | <a href="https://static.igem.org/mediawiki/2013/2/2b/UNIK_Copenhagen_Notebook_ref_16.35.jpg" target="_blank"> <img src="https://static.igem.org/mediawiki/2013/2/2b/UNIK_Copenhagen_Notebook_ref_16.35.jpg" width="200"></a><br> |
<p> Gel no. 3: Colony PCR of eFbFP-pDONR207 and MamC-eGFP-pDONR207 </p> | <p> Gel no. 3: Colony PCR of eFbFP-pDONR207 and MamC-eGFP-pDONR207 </p> | ||
</div> | </div> | ||
| - | <div id=" | + | <div id="under_page2"> |
| - | <img src="https://static.igem.org/mediawiki/2013/b/b5/UNIK_Copenhagen_Notebook_ref_16.45.jpg" | + | <a href="https://static.igem.org/mediawiki/2013/b/b5/UNIK_Copenhagen_Notebook_ref_16.45.jpg" target="_blank"><img src="https://static.igem.org/mediawiki/2013/b/b5/UNIK_Copenhagen_Notebook_ref_16.45.jpg" height="200"> </a> |
<p> Gel no. 4: 16S PCR amplification. Negative control - <i>H. C. Ørsteds Parken</i> sample - <i>Christiania</i> sample </p> | <p> Gel no. 4: 16S PCR amplification. Negative control - <i>H. C. Ørsteds Parken</i> sample - <i>Christiania</i> sample </p> | ||
</div> | </div> | ||
| Line 366: | Line 400: | ||
<div class="right_page"> | <div class="right_page"> | ||
<p> | <p> | ||
| - | Gateway BP reaction was performed to clone MamC-eGFP into pDONR207 using MamC-eGFP (GTW PCR product) no. 3 and 4 was used and eFbFP (GTW PCR product) no. 3 and 7. Colony PCR was carried out ( | + | Gateway BP reaction was performed to clone MamC-eGFP into pDONR207 using MamC-eGFP (GTW PCR product) no. 3 and 4 was used and eFbFP (GTW PCR product) no. 3 and 7. Colony PCR was carried out (see gel no. 3). |
<br> | <br> | ||
Colonies 2, 3, 5, 7, 9, 11, 13, 14 were selected to make liquid culture. Miniprep was done and samples were sent for sequencing. | Colonies 2, 3, 5, 7, 9, 11, 13, 14 were selected to make liquid culture. Miniprep was done and samples were sent for sequencing. | ||
| Line 382: | Line 416: | ||
<u>The CPH strain</u> | <u>The CPH strain</u> | ||
<br> | <br> | ||
| - | PCR amplification of 16S from our lake sample (enriched) and run on gel ( | + | PCR amplification of 16S from our lake sample (enriched) and run on gel (see gel no. 4). |
The gel shows some amount of contamination in the negative control (no template). | The gel shows some amount of contamination in the negative control (no template). | ||
| Line 533: | Line 567: | ||
<br> | <br> | ||
Confocal images were taken of MamC-eGFP-pJAM1786 and eGFP-pBBR1MCS-2 alongside MS-1 and MSR-1 cultures. | Confocal images were taken of MamC-eGFP-pJAM1786 and eGFP-pBBR1MCS-2 alongside MS-1 and MSR-1 cultures. | ||
| - | <br><br> | + | <br>Hover the mouse over the picture to see!<br></p> |
| + | <div id="confocals"> | ||
| + | <div id="confocal"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/7/7a/UNIK_Copenhagen_confocal_image4.jpg" width="160px"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/e/e6/UNIK_Copenhagen_confocal_image3.jpg" width="160px"> | ||
| + | </div> | ||
| + | |||
| + | <div id="confocal"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/f/f4/UNIK_Copenhagen_confocal_image2.jpg" width="160px"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/0/02/UNIK_Copenhagen_confocal_image1.jpg" width="160px"> | ||
| + | </div> | ||
| + | </div> | ||
| + | <p> First photo: eGFP, Second photo: MamC-eGFP </p> | ||
| + | |||
| - | |||
</div> | </div> | ||
</div> | </div> | ||
| Line 544: | Line 590: | ||
<div class="left_page"> | <div class="left_page"> | ||
<h1>Week 12 - September 23<sup>rd</sup>-29<sup>th</sup></h1> | <h1>Week 12 - September 23<sup>rd</sup>-29<sup>th</sup></h1> | ||
| - | <p> | + | <p> |
| + | <u>Assembly of constructs</u><br> | ||
| + | Miniprep was carried out on the frozen pellets from last week. LR reaction was setup to yield eGFP-pJAM1786 and eFbFP-pJAM1786. Colony PCR was performed. | ||
| + | <br> | ||
| + | <a href="https://static.igem.org/mediawiki/2013/8/88/UNIK_Copenhagen_Notebook_ref_0109.jpg" target="_blank"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/8/88/UNIK_Copenhagen_Notebook_ref_0109.jpg" height="120"> | ||
| + | </a> | ||
| + | <br><br> | ||
| + | Looking at the bands from the Colony PCR, one can tell that there might have been problems with the gel. We decided to consider the odd looking bands in the left to be due to the gel and therefore we inoculated the restreaked cultures into liquid LB. | ||
| + | <br><br> | ||
| + | Using an opaque ELISA reader plate we measured fluorescence of 7 eGFP-pJAM1786 and 6 eFbFP-pJAM1786 cultures | ||
| + | </p> | ||
</div> | </div> | ||
<div class="right_page"> | <div class="right_page"> | ||
| - | <p> | + | <p> |
| + | (3 control cultures, containing pBBR1MCS-2 without insert, were used as well). | ||
| + | The measurements failed to show eFbFP-pJAM1786 to be significantly different from the control cultures. This was in contrast to eGFP-pJAM1786 that was shown be fluorescent. | ||
| + | <br><br> | ||
| + | Western blot was performed on the 3 strains above (eFbFP, eGFP and control), MamC-eGFP-pJAM1786 and a GFP control. Strangly, the MamC-eGFP-pJAM1786 cultures did not show fluorescence. This was highly unexpected since the cultures that was used for this glycerol stock earlier had been shown to be fluorescent. Therefore, we did not expect any bands in the MamC-eGFP lanes. | ||
| + | |||
| + | The western blot somewhat failed since only one of the standard protein markers showed in the membrane. (See a visualization <a href="https://static.igem.org/mediawiki/2013/8/83/UNIK_Copenhagen_Notebook_ref_Western.jpg | ||
| + | " target="_blank">here</a>). The GFP standard (band at the right side) seem to be highly overloaded. The lanes to left is eGFP-pJAM in different amounts. | ||
| + | |||
| + | |||
| + | |||
| + | </p> | ||
</div> | </div> | ||
</div> | </div> | ||
| + | |||
| + | |||
| + | <div id="content13" class="content"> | ||
| + | <div class="left_page"> | ||
| + | <h1>Week 12 - Sept. 30<sup>th</sup>-Oct. 6<sup>th</sup></h1> | ||
| + | <p> | ||
| + | <u> Wrapping it up - and preparing for Lyon </u> | ||
| + | <br> | ||
| + | This last week before the jamboree we've spent finishing up our parts characterisation and getting the wiki shining before the freeze. | ||
| + | <br><br> | ||
| + | The presentation crew have been working on their performance for Lyon. The rest of the team and supervisors got to see a rehersal and feed them inputs and comments. We're all looking forward to put on a show in Lyon. | ||
| + | |||
| + | <br><br> | ||
| + | Team Magneto is ready and looking forward to a nice time in Lyon! | ||
| + | |||
| + | |||
| + | </div> | ||
| + | <div class="right_page"> | ||
| + | |||
| + | <div id="middle"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/0/01/EuJamLogoiGemLyon.png" width="250"> | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
</div> | </div> | ||
| Line 566: | Line 659: | ||
<li><a href="javascript:ThisTab('tab11', 'content11');" id="tab11">11</a></li> | <li><a href="javascript:ThisTab('tab11', 'content11');" id="tab11">11</a></li> | ||
<li><a href="javascript:ThisTab('tab12', 'content12');" id="tab12">12</a></li> | <li><a href="javascript:ThisTab('tab12', 'content12');" id="tab12">12</a></li> | ||
| + | <li><a href="javascript:ThisTab('tab13', 'content13');" id="tab13">13</a></li> | ||
</ul> | </ul> | ||
</div> | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| - | |||
| - | |||
</html> | </html> | ||
Latest revision as of 22:15, 3 October 2013
Notebook
Get our Nootebook in a printable version here.
If nothing happens when you click, check if you have disabled popup-windows.
For more detailed descriptions of the methods used, please see our protocols.
Week 1 - July 8th-14th
Culturing of magnetotactic bacteria (MTBs)
Making base medium for growing magnetotactic bacteria. This medium was also used to make agar plates.
MS-1 inoculated in liquid base medium. Falcon tubes were flushed with the nitrogen before use and incubated at 28 degrees.
Plates were kept in a closed container with an Oxoid AnaeroGen pad.
Assembly of constructs
Glycerol stock of E. coli with pBBR1MCS-2 was thawed to grow on these plates.
Primers for MamC (MS-1 strain) was ordered.
Week 3 - July 22nd-28th
Culturing of MTBs
making charcoal media and plates. MS-1 and MSR-1 was inoculated. Liquid cultures were setup in Hungate tubes 200ul, 400ul and 600ul of each strain (MS-1 and MSR-1). Charcoal plates were inoculated as well with both strains.
Assembly of constructs
Colony PCRs of eFbFP-pEX-A and eGFP-pDRIVE. Only eFbFP was successful.

Overnight cultures were made of colonies 1-4 (eFbFP-pEX-A). Miniprep of eFbFP-pEX-A
cultures. Samples sent for sequencing.
Overnight cultures were made of colonies 1-4 (eFbFP-pEX-A).
Miniprep of eFbFP-pEX-A cultures. Samples sent for sequencing.
Colony PCR of eGFP-pDRIVE was repeated with the same outcome.
Cloning and transformation was also repeated. CloneJET PCR cloning kit was used to create eGFP-pJET (pJET was chosen instead of pDRIVE). Not successful.
Gradient PCR of eGFP premade vector (56-65 degrees). Gel electrophoresis was carried out and the bands were purified from the gel.
An attempt to clone eGFP into pDRIVE was not successful.
The cloning of eFbFP into expression vector pBBR1MCS-2 was successful instead.
Colony PCR of eGFP-pDRIVE and eFbFP-pBBR1MCS-2. Gel electrophoresis revealed only one colony to be successful (eFbFP-pBBR1MCS-2 no. 17).
Week 4 - July 29th- August 4th
Assembly of constructs
Colony PCR of eGFP-pDRIVE was repeated with increased elongation time (1 min.)
 Cloning looks successful and colonies 1-4 were set up as liquid cultures overnight.
Cloning looks successful and colonies 1-4 were set up as liquid cultures overnight.
Colony PCR of eFbFP-pBBR1MCS-2 because only one colony was positive in week 3. Increased elongation time.
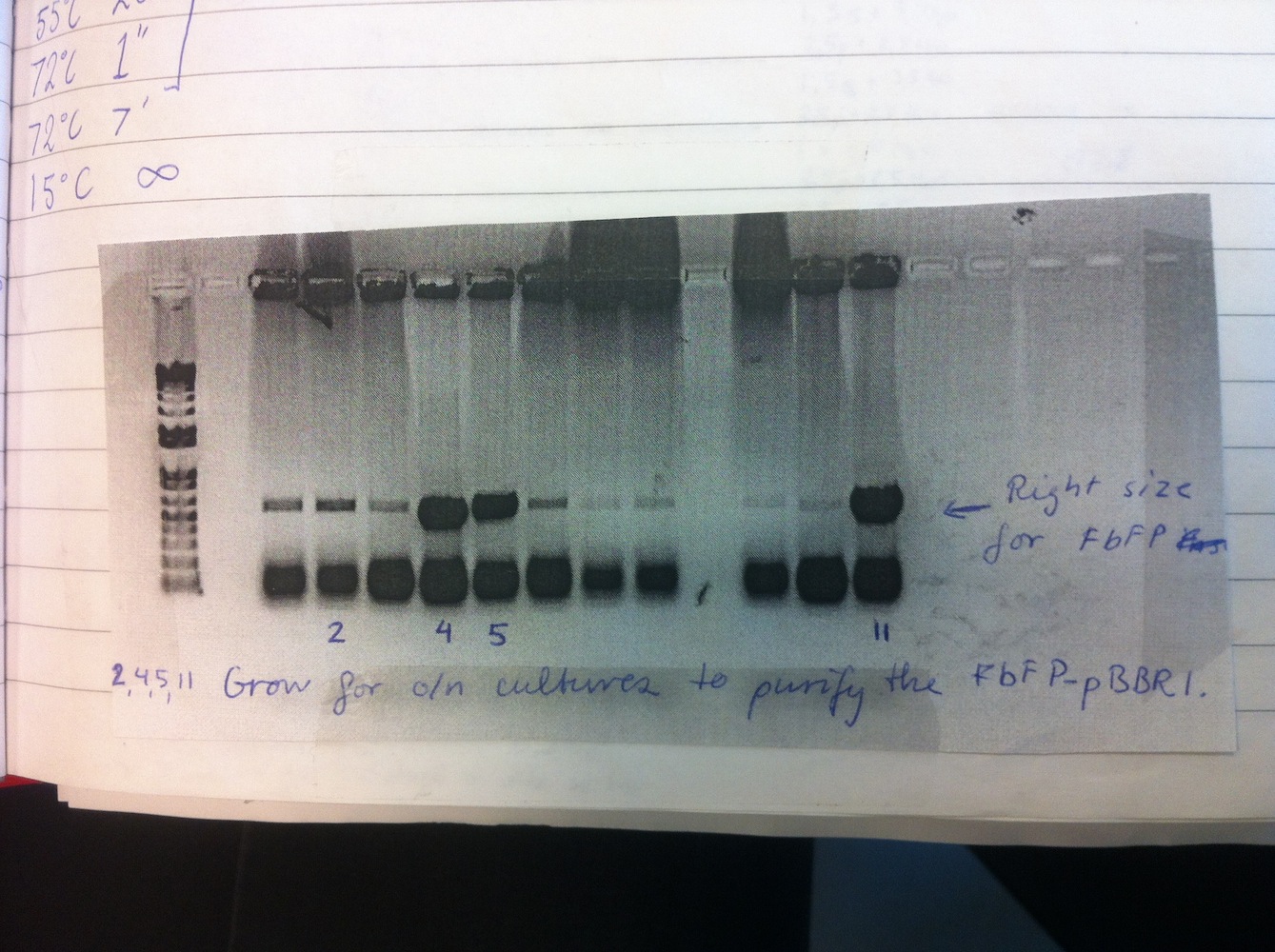
Overnight liquid cultures were setup for colonies 2, 4, 5 and 11.
Miniprep was performed on these cultures alongside eGFP-pDRIVE colonies 1-4. This gave good yields and the plasmids were sent for sequencing. This showed eGFP-pDRIVE no. 4 and eFbFP-pBBR1MCS-2 no. 17 was correct!
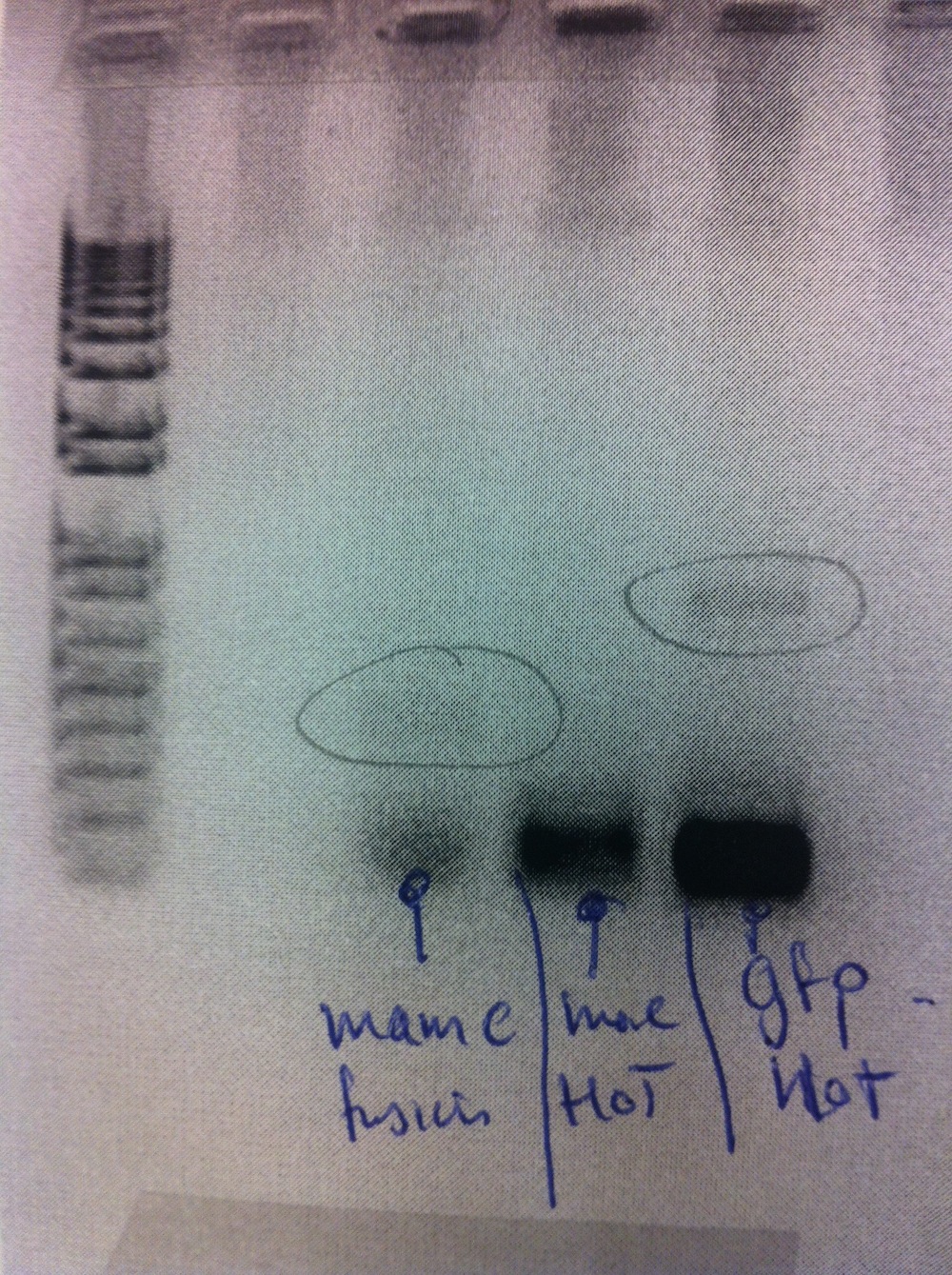
Cloning and transformation of eGFP-pBBR1MCS-2 using plasmid no. 4 (mentioned just above). The transformation only gave rise to 2 colonies, thus suggesting the cloning to be unsuccessful. The cloning was repeated and transformation was carried out again. Colony PCR was performed alongside MamC-pJET (see gel below).
Liquid cultures were setup from eFbFP-pBBR1MCS-2 no. 2, 4, 5, 11, 17 for 2 hours in order to measure fluorescence. First, OD(600nm) was measured to ensure equal density of cells. The measurements did not show fluorescence (ex. 450 nm, em. 495 nm) from the cultures.
Cloning and transformation of MamC-pDRIVE. Colony PCR showed the cloning to be of cryptic success. Colony PCR was redone without success.
Cloning was repeated using the CloneJET PCR cloning kit. MamC-pJET was transformed and plated. Colony PCR was carried out alongside eGFP-pBBR1MCS-2.
 The gel electrophoresis indicated MamC-pJET to be successfully cloned. However, only a few of the eGFP-pBBR1MCS-2 colonies appeared to have contain the insert.
The gel electrophoresis indicated MamC-pJET to be successfully cloned. However, only a few of the eGFP-pBBR1MCS-2 colonies appeared to have contain the insert.
Week 5 - August 5th-11th
Culturing of MTBs
Colony PCR using MamC primer was done with MS-1 and MSR-1 colonies. MamC was amplified from the genomic DNA. This did not give rise to any bands via gel electrophoresis.
MamC was, however, successfully amplified from MSR-1 later in the week (see gel below).

Assembly of constructs
Colonies 1, 2, 3 (MamC-pJET) and II, IV, VIII (eGFP-pBBR1MCS-2) were setup in liquid cultures (see colony PCR from previous week). Miniprep was carried out and plasmids were sent for sequencing. All MamC sequences were perfect while all eGFP sequences were wrong. The cloning of eGFP into pDRIVE was successfully redone (documentation lost). Fluorescence was also measured of eFbFP-pBBR1MCS-2 strains. No fluorescence detected as the sequencing explains. Inducing with IPTG (final conc. 1 mM) had no effect.
The CPH strain
Enrichment of magnetotactic bacteria was done according to the protocol. It was unclear whether the bacteria observed in the microscope were actually magnetotactic. They were compared to MS-1 and MSR-1 samples.
Week 6 - August 12th-18th
Assembly of constructs
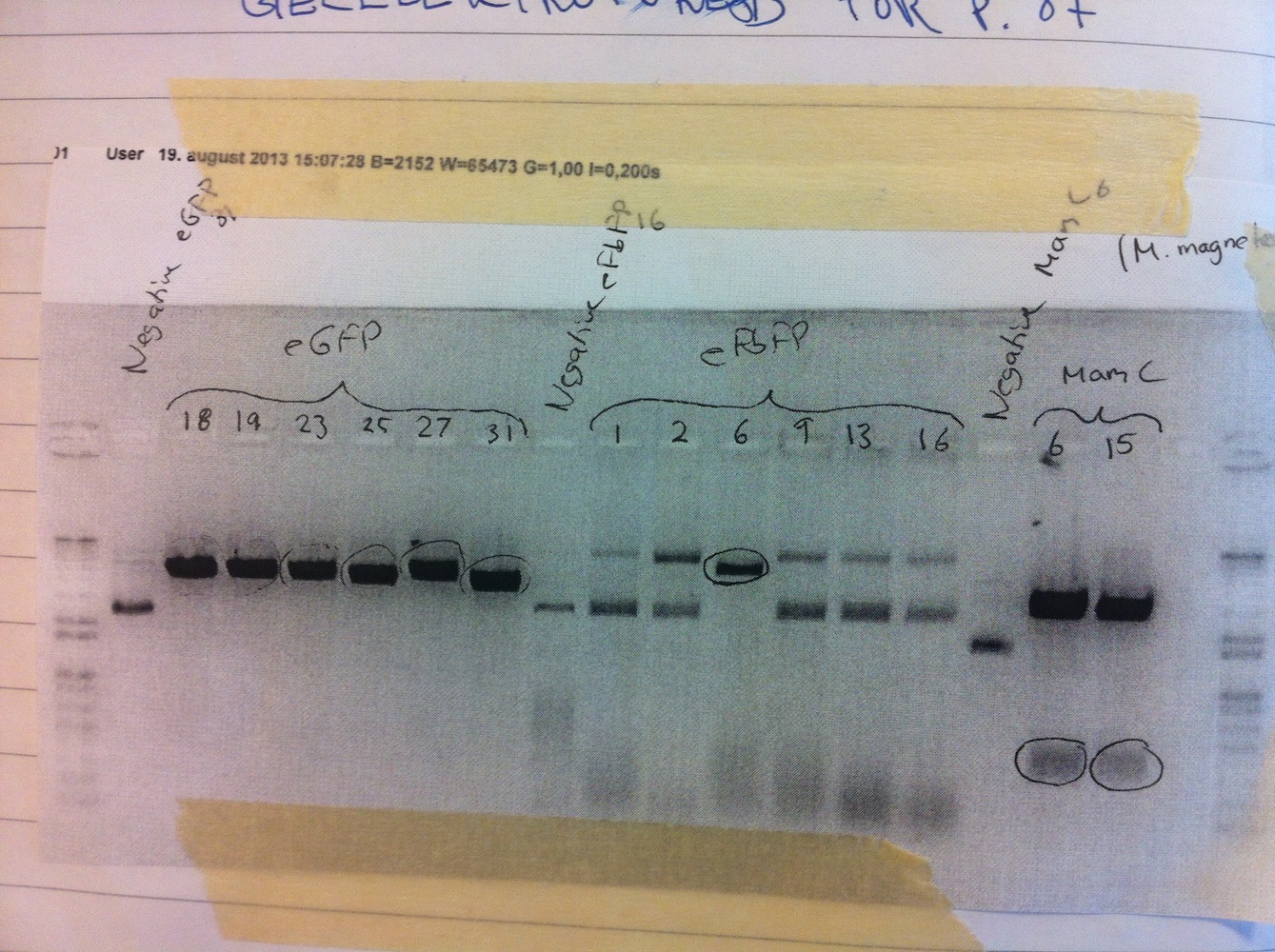
Since we couldn't detect any fluorescence from eFbFP-pBBR1MCS-2 we needed troubleshoot this. We found a frameshift in the vector and thus we repeated the cloning of eFbFP into pBBR1MCS-2. This was done alongside cloning eGFP into pBBR1MCS-2. Transformation and colony PCR were carried out. All colonies appeared to be positive on the gel electrophoresis (see picture below).

MamC-pJET did not contain the restriction sites we needed. Therefore, the whole procedure for obtaining MamC-pJET was repeated using other primers (with different overhangs).
Six of each genotype were set up for liquid cultures overnight (colonies 1, 2, 6, 9, 13, 16 for eFbFP and 18, 19, 23, 25, 27, 31 for eGFP). Miniprep was done and samples were sent for sequencing. None of the eFbFP gave rise to good quality sequencing. Colonies 18, 19, 23 and 27 of eGFP contained the correct sequence.
Empty pBBR1MCS-2 vector was transformed into E. cloni in order to obtain more stock vector.
Culturing of MTBs
MSR-1 base medium culture was inoculated in new medium and put in 28 degrees shaker over the weekend. This was done to check whether the strain would grow aerobically as well.
Week 7 - August 19th-25th
Assembly of constructs
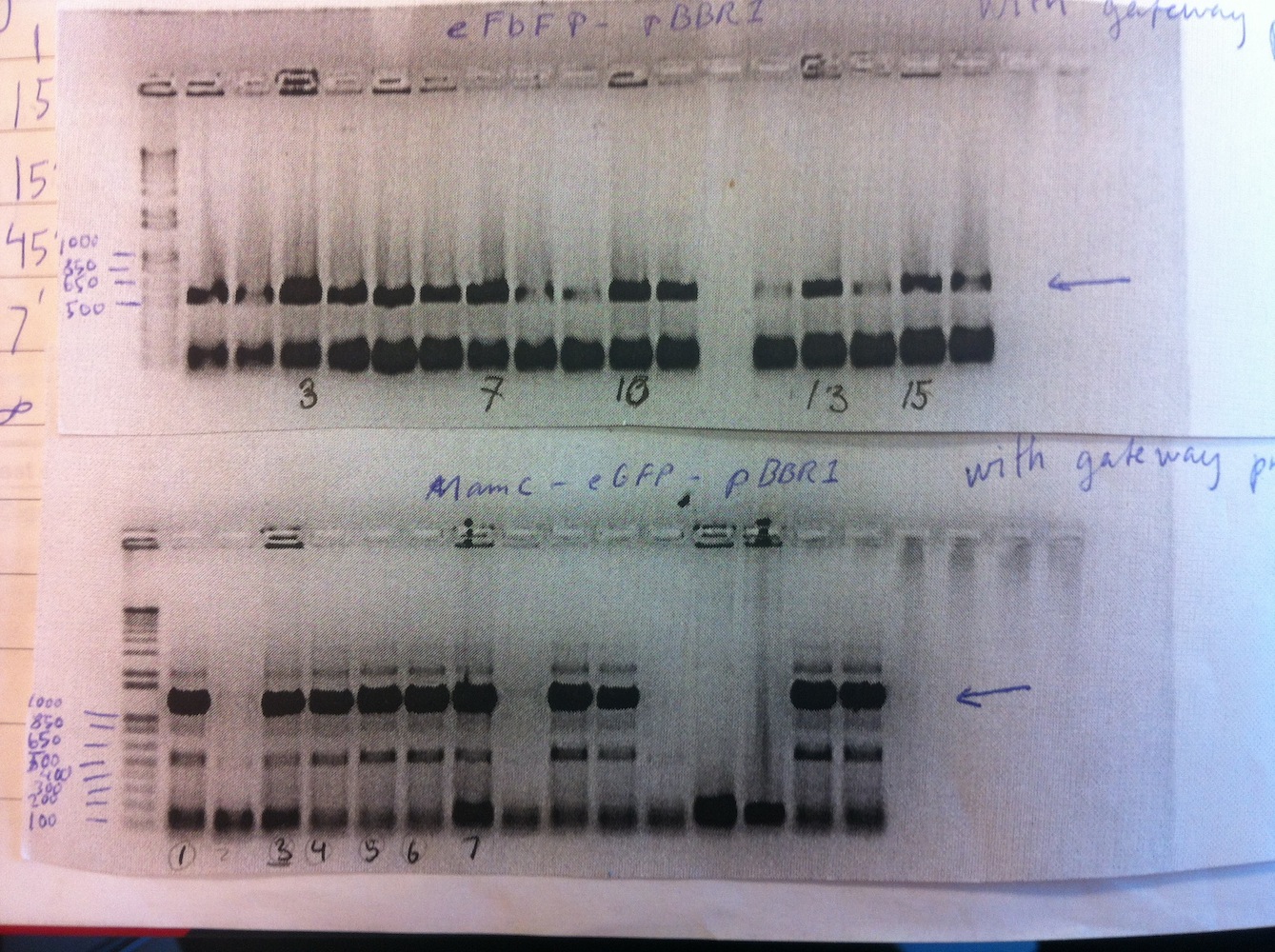
Plasmids from last week eFbFP/eGFP-pBBR1MCS-2 were digested alongside the newly assembled MamC-pJET in order to assess whether the restriction sites were present or not (see gel no. 1).
All lanes with eGFP or MamC were positive (see gel above) and the bands were purified from the gel. From this purification very poor concentration was obtained. From the lanes with eFbFP, no. 6 stands out from the negative control. Therefore we decided we continue working with this plasmid. A re-ligation of eFbFP-pBBR1MCS-2 was performed.
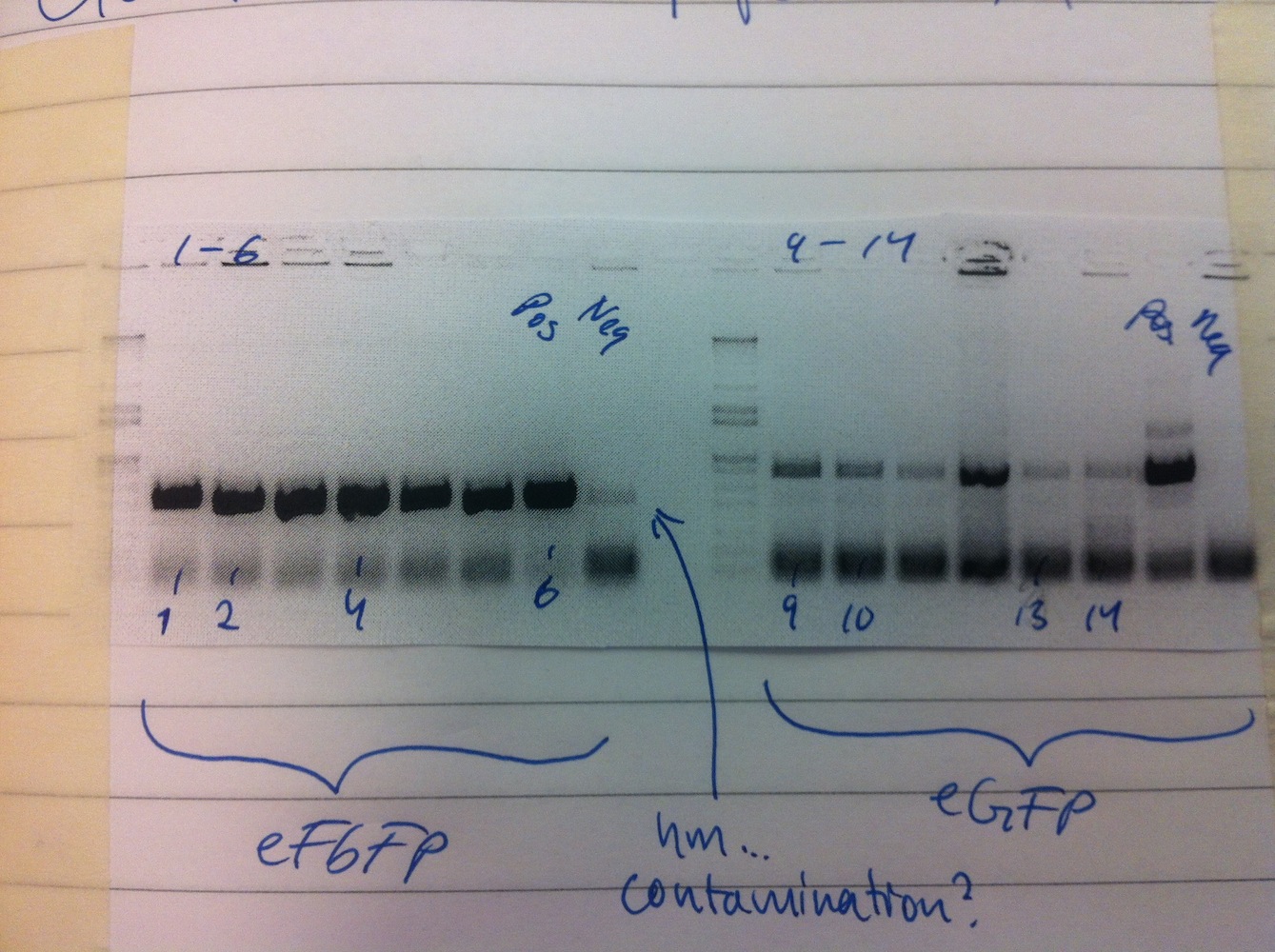
Cloning of MamC into linearized eGFP-pBBR1MCS-2 and subsequently transforming the plasmid was also done. Colony PCR was carried out on MamC-eGFP-pBBR1MCS-2 and eFbFP-pBBR1MCS-2. We decided to use the Gateway (GTW) system to express our constructs (see gel no. 2).
To save time the colony PCR was performed using gateway primers (MamC F and eGFP R, eFbFP F and R). The PCR product of MamC-eGFP-pBBR1MCS-2 colonies 1, 3, 4, 5, 6 and eFbFP-pBBR1MCS-2 3, 7, 10, 13, 15 were PCR purified.
Gateway BP reaction was performed to clone MamC-eGFP into pDONR207 using MamC-eGFP (GTW PCR product) no. 3 and 4 was used and eFbFP (GTW PCR product) no. 3 and 7. Colony PCR was carried out (see gel no. 3).
Colonies 2, 3, 5, 7, 9, 11, 13, 14 were selected to make liquid culture. Miniprep was done and samples were sent for sequencing.
BioBrick submission
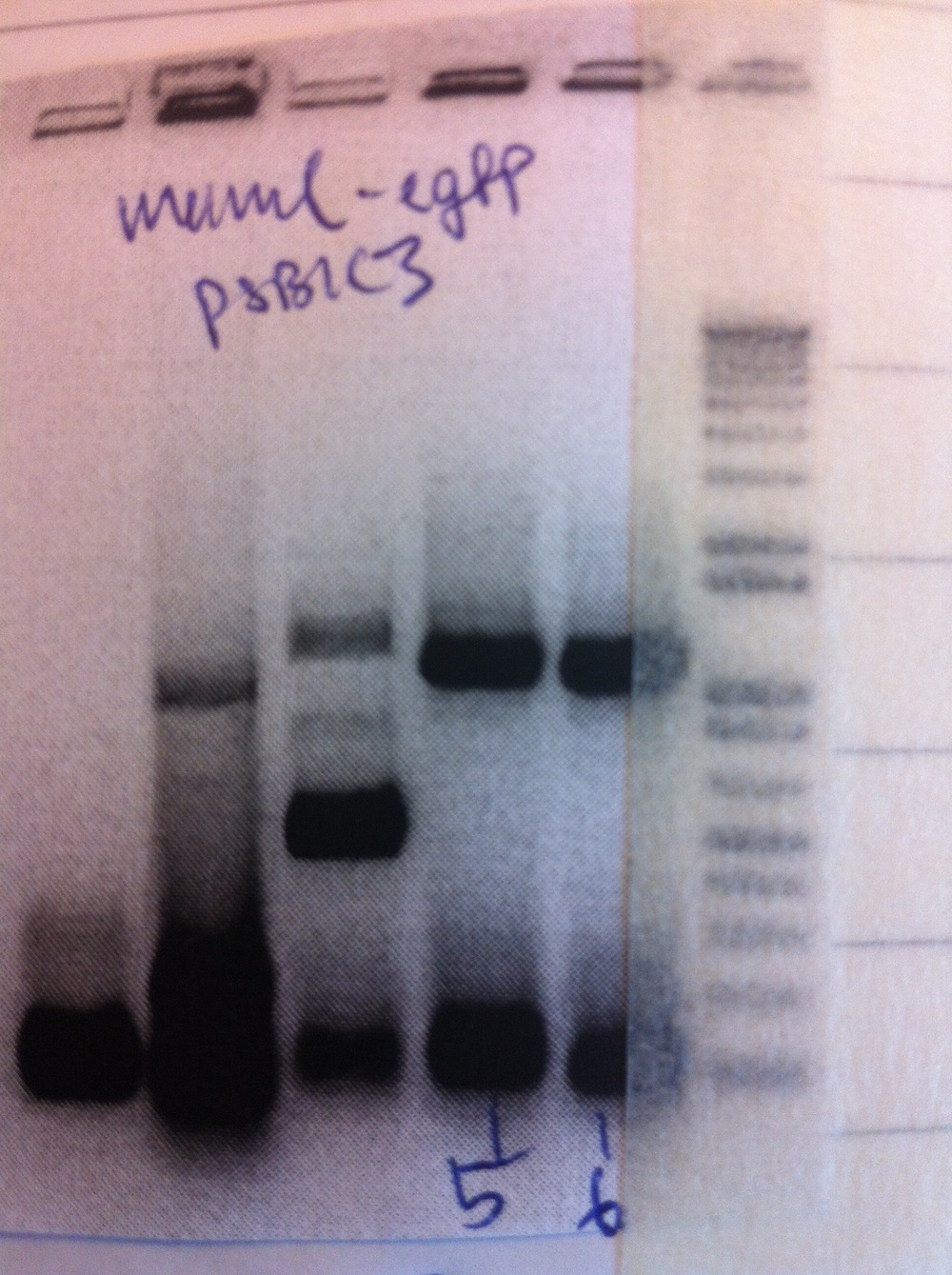
PCR amplification of MamC15 (week 6), eFbFP-pEX-A, eGFP18 (week 6) and eGFP19 (week 6). Products were run on a gel and the bands were purified. Ligation was set up to clone the fragments into pSB1C3 and subsequently transformation was performed as well.
Culturing of MTBs
Microscopy of MSR-1 in order to check morphology is corresponding to what is reported in the literature (Jogler & Schüler, Annu. Rev. Microbiol. 2009. 63:501-21). We did not see any signs of bacterial growth in the liquid cultures set up in week 6.
The CPH strain
PCR amplification of 16S from our lake sample (enriched) and run on gel (see gel no. 4).
The gel shows some amount of contamination in the negative control (no template).
Week 8 - August 26th-September 1st
Assembly of constructs
Sequencing of eFbFP-pDONR207 and MamC-eGFP-pDONR207. Gateway LR reaction for cloning MamC-eGFP (no 11 and no 13) into pJAM1786 and transformation into E. coli.
Colony PCR was performed on MamC-eGFP-pJAM1786 and eFbFP-pDONR207 (see gel below)

Colonies 1, 2, 6, 10, 11, 13, 14, 16 were inoculated in liquid culture and grown overnight. Miniprep was performed and LR reaction was done using eFbFP-pDONR207 no 11. Enhanced FbFP-pJAM1786 was transformed and grown overnight.
Expressing eFbFP in the gateway system was repeated. PCR amplification using Gateway primers
on eFbFP-pEX-A was carried out. Gateway BP reaction was done to move the fragment into pDONR207. Afterwards the plasmid was transformed into E. coli.
The CPH strain
Cloning of 16S PCR products into pJET and subsequent transformation into E. coli. Liquid cultures were grown inoculating cells from 24 different colonies (12 from H.C. Ørsted Parken sample and 12 from Christiania sample). Miniprep was carried out and samples were sent for sequencing.
BioBrick submission
Colony PCR was performed.
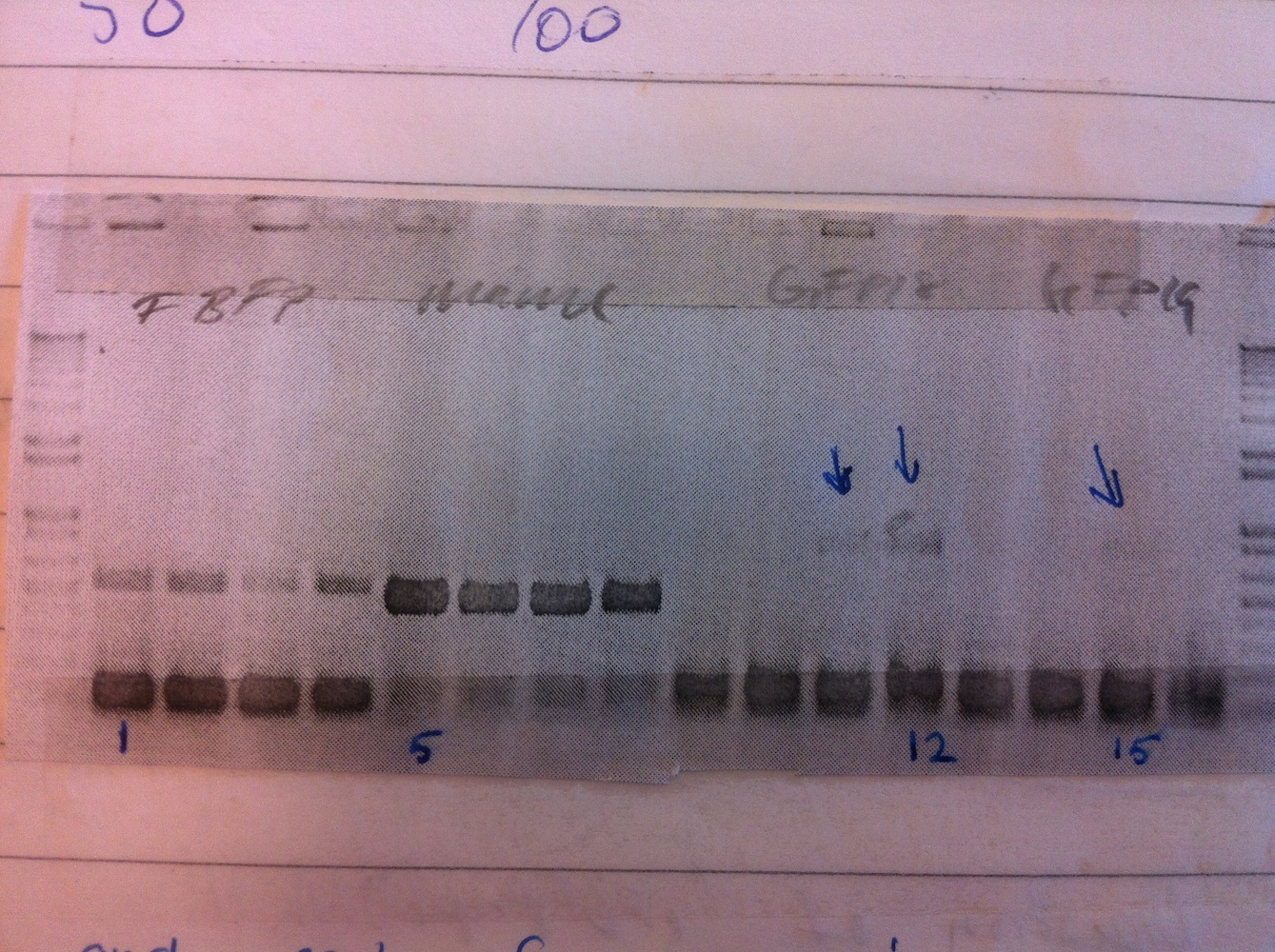 Colonies 1, 5, 12 and 15 were inoculated to make liquid cultures, that were grown overnight.
Colonies 1, 5, 12 and 15 were inoculated to make liquid cultures, that were grown overnight.
Week 9 - September 2nd-8th
Assembly of constructs
Cultures of eFbFP-pJAM1786 was minipreped and samples were sent for sequencing. Microscopy revealed MamC-eGFP-pJAM1786 and eFbFP-pJAM1786 emitted fluorescence. To measure fluorescence quantitatively, we set up 8 liquid cultures of MamC-eGFP-pJAM1786 and 3 of MamC-pSB1C3 as a non-fluorescent control. Fluorescence was measured for dilutions 1X, 2X, 5X, 10X, 50X (Ex. 485 nm em. 535 nm.) using an ELISA reader (black plate) and gave rise characterization for BioBrick BBa_K1094401.
OD(600nm) was also measured to ensure approx. equal cell density.
BioBrick submission
Miniprep was performed and samples sent for sequencing. The sequencing results revealed that sequences of MamC-pSB1C3, eGFP(18)-pSB1C3
and eGFP(19)-pSB1C3 were correct while eFbFP-pSB1C3 held the wrong insert. We uncovered that this was due two "illegal" restriction sites in the FbFP sequence. We, therefore, ordered another synthetic gene from IDT with the restriction sites removed.
To submit MamC-eGFP as a BioBrick we PCR amplified (1:20 min elongation time) the whole construct (MamC F primer and eGFP R primer). To do this we used 3 different templates.
Afterwards, digestion and ligation into pSB1C3 was performed and the plasmid was transformed into E. coli.
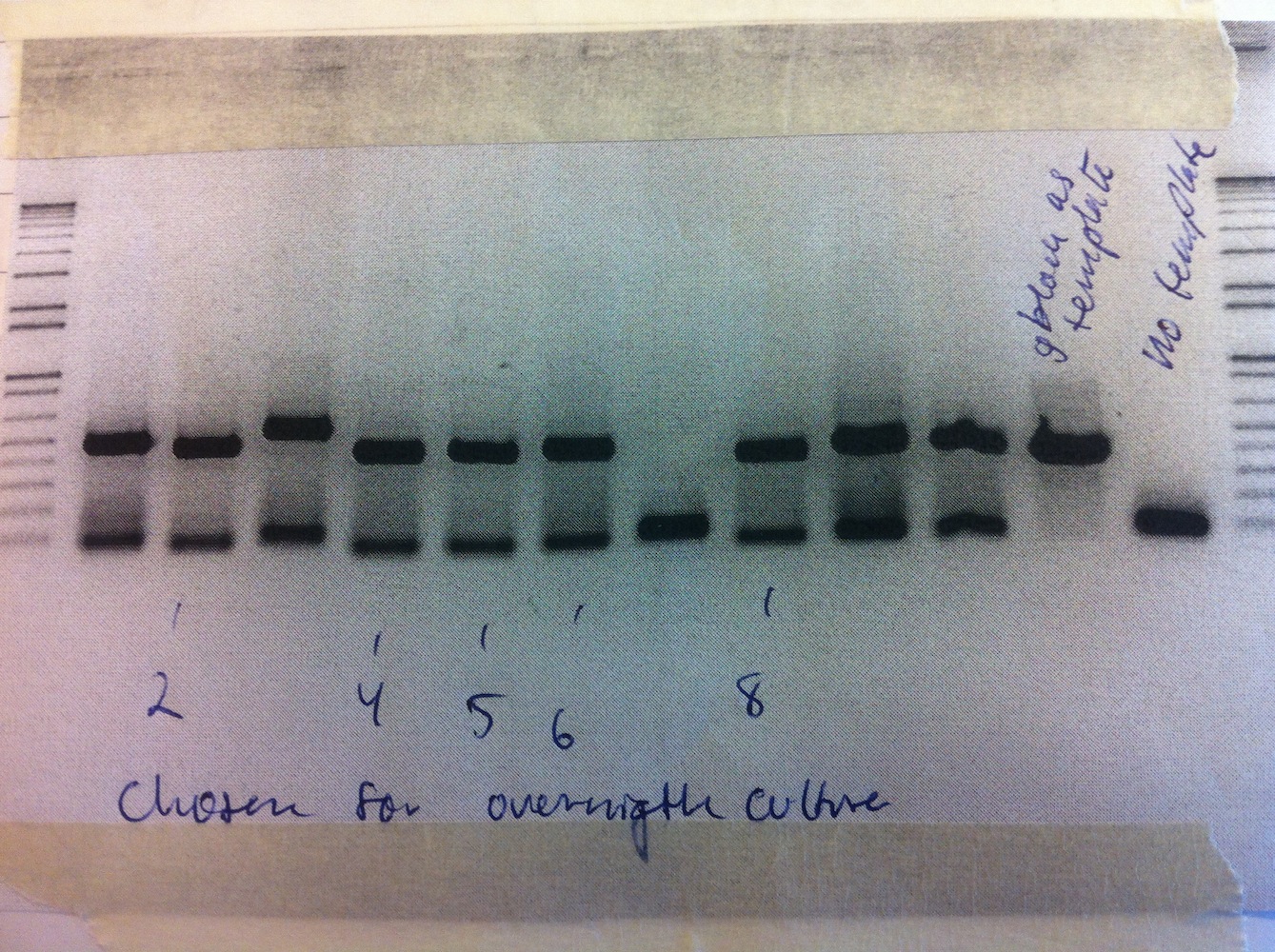
Colony PCR was performed.
Colonies 5 and 6 were inoculated into liquid culture and grown overnight. Miniprep was performed and samples sent for sequencing.
Week 10 - September 9th-15th
Assembly of constructs
The newly synthesized eFbFP came in. Traditional cloning was used to put it into pSB1C3 and the plasmid was transformed. Colony PCR was performed.
Colonies 2, 4, 5, 6, and 8 were inoculated in liquid culture. Miniprep was carried out and samples sent for sequencing.
BioBrick submission
We sent 4 BioBricks for the registry. Enhanced FbFP-pSB1C3 was sent in 5 copies because we did not have the sequencing results yet.
Culturing of MTBs
New base media was made according to Schüler et al. 1991 (see protocols). A variant was made with 0,2% agar as well. Both MS-1 and MSR-1 were inoculated and grown in 28 degrees in both media. 3 days after the tubes showed signs of bacteria growing.
Confocal imaging
Cultures set up for confocal imaging. Strains MamC-eGFP-pJAM1786 and eGFP-pBBR1MCS-2 were inoculated.
Week 11 - September 16th-22nd
Assembly of constructs
To express and characterize eFbFP and eGFP (without MamC) we needed to move the sequences to the Gateway system. Therefore, PCR using Gateway primers was performed and a BP reaction was set up in order to clone eFbFP (new) and eGFP into pDONR207. Transformation and colony PCR was performed.
Colonies 1, 2, 4, 6, 9, 10, 13, 14 were put into liquid culture and grown overnight. Cultures were centrifuged after 1 day, medium was removed and the cells were stored at -20 degrees.
Confocal imaging
Confocal images were taken of MamC-eGFP-pJAM1786 and eGFP-pBBR1MCS-2 alongside MS-1 and MSR-1 cultures.
Hover the mouse over the picture to see!




First photo: eGFP, Second photo: MamC-eGFP
Week 12 - September 23rd-29th
Assembly of constructs
Miniprep was carried out on the frozen pellets from last week. LR reaction was setup to yield eGFP-pJAM1786 and eFbFP-pJAM1786. Colony PCR was performed.

Looking at the bands from the Colony PCR, one can tell that there might have been problems with the gel. We decided to consider the odd looking bands in the left to be due to the gel and therefore we inoculated the restreaked cultures into liquid LB.
Using an opaque ELISA reader plate we measured fluorescence of 7 eGFP-pJAM1786 and 6 eFbFP-pJAM1786 cultures
(3 control cultures, containing pBBR1MCS-2 without insert, were used as well).
The measurements failed to show eFbFP-pJAM1786 to be significantly different from the control cultures. This was in contrast to eGFP-pJAM1786 that was shown be fluorescent.
Western blot was performed on the 3 strains above (eFbFP, eGFP and control), MamC-eGFP-pJAM1786 and a GFP control. Strangly, the MamC-eGFP-pJAM1786 cultures did not show fluorescence. This was highly unexpected since the cultures that was used for this glycerol stock earlier had been shown to be fluorescent. Therefore, we did not expect any bands in the MamC-eGFP lanes.
The western blot somewhat failed since only one of the standard protein markers showed in the membrane. (See a visualization here). The GFP standard (band at the right side) seem to be highly overloaded. The lanes to left is eGFP-pJAM in different amounts.
Week 12 - Sept. 30th-Oct. 6th
Wrapping it up - and preparing for Lyon
This last week before the jamboree we've spent finishing up our parts characterisation and getting the wiki shining before the freeze.
The presentation crew have been working on their performance for Lyon. The rest of the team and supervisors got to see a rehersal and feed them inputs and comments. We're all looking forward to put on a show in Lyon.
Team Magneto is ready and looking forward to a nice time in Lyon!

 "
"