Team:DTU-Denmark/Notebook/13 August 2013
From 2013.igem.org
(→PCR (promoter library)) |
(→SLP screening PCR) |
||
| (12 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:DTU-Denmark/Templates/StartPage|13 August 2013}} | {{:Team:DTU-Denmark/Templates/StartPage|13 August 2013}} | ||
| - | + | Navigate to the [[Team:DTU-Denmark/Notebook/12_August_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/14_August_2013|Next]] Entry | |
| - | = | + | =Lab 208= |
<hr/> | <hr/> | ||
==Main purpose== | ==Main purpose== | ||
<hr/> | <hr/> | ||
| + | *PCR to amplify USER fragments of the constitutive SPL; | ||
| + | *PCR of Nir2 with USER primers; | ||
| + | *Gel purification of araBAD SPL. | ||
==Who was in the lab== | ==Who was in the lab== | ||
| Line 19: | Line 22: | ||
Used 5% DMSO, 2 uL of mM MgCl2 and GC-buffer. | Used 5% DMSO, 2 uL of mM MgCl2 and GC-buffer. | ||
| - | constitutive SPL | + | <u>constitutive SPL</u> |
*template: pZA21::RFP | *template: pZA21::RFP | ||
*primers: 52a, 52b1 | *primers: 52a, 52b1 | ||
| Line 25: | Line 28: | ||
*elongation time: 3:00 | *elongation time: 3:00 | ||
| - | constitutive reference | + | <u>constitutive reference</u> |
*template: pZA21::RFP | *template: pZA21::RFP | ||
*primers: 52a, 52b2 | *primers: 52a, 52b2 | ||
| Line 33: | Line 36: | ||
Wanted to make negative for the two above reactions but accidentally put the template in the master mix. | Wanted to make negative for the two above reactions but accidentally put the template in the master mix. | ||
| - | arabinose reference | + | <u>arabinose reference</u> |
*template: pZA21:araBAD:RFP (labeled Ara) | *template: pZA21:araBAD:RFP (labeled Ara) | ||
*primers: 51a,51b1 | *primers: 51a,51b1 | ||
| Line 39: | Line 42: | ||
*elongation time: 4:00 | *elongation time: 4:00 | ||
| - | program (used for all 3 reactions) | + | program (used for all 3 reactions): |
{| class="wikitable" style="text-align: right" | {| class="wikitable" style="text-align: right" | ||
! temperature !! time !! cycles | ! temperature !! time !! cycles | ||
| Line 57: | Line 60: | ||
|} | |} | ||
| - | ===PCR Nir2=== | + | ===PCR of Nir2 with USER primers=== |
| - | |||
{| class="wikitable" style="text-align: right" | {| class="wikitable" style="text-align: right" | ||
! sample nr. !! buffer !! %DMSO !! program | ! sample nr. !! buffer !! %DMSO !! program | ||
|- | |- | ||
| - | | 1 || | + | | 1 || GC || 2 || 55C |
|- | |- | ||
| - | | 2 || | + | | 2 ||GC || 3 || 55C |
|- | |- | ||
| - | | 3 || | + | | 3 || GC || 5 || 55C |
|- | |- | ||
| - | | 4 || || | + | | 4 || HF|| 2 || 55C |
|- | |- | ||
| - | | 5 || | + | | 5 || HF || 3 || 55C |
|- | |- | ||
| - | | 6 || | + | | 6 || HF || 5 || 55C |
|- | |- | ||
| - | | 7 || | + | | 7 || GC || 2 || ramp |
|- | |- | ||
| - | | 8 || | + | | 8 || GC || 3 || ramp |
|- | |- | ||
| - | | 9 || | + | | 9 || GC || 5 || ramp |
|- | |- | ||
| - | | 10 || | + | | 10 || HF || 2 || ramp |
|- | |- | ||
| - | | 11 || | + | | 11 || HF || 3 || ramp |
|- | |- | ||
| - | | 12 || | + | | 12 || HF || 5 || ramp |
|- | |- | ||
|} | |} | ||
| + | |||
| + | * PCR mix according to standard protocol with changes: addition of DMSO in 3 different final concentrations (2%, 3%, 5%); two different buffers (HF 5x and GC 5x), amount of added water was dependent on volume of added DMSO. | ||
| + | * Primers for Nir2 - 40a, 40b | ||
| + | * Template - fragment Nir2 amplified with non-uracil primers, 3uL were used | ||
| + | * Polymerase x7 | ||
| + | * Program A99, samles 1-6, was based on standard PCR program with 55 C and 1 min of annealing parameters and 5 min of extension time. | ||
| + | * Program A1, samples 7-12, PCR with initial annealing temperature of 59C falling down to 53C (rate 0.1 C/min). 5 min of extension time. | ||
===Gel purification=== | ===Gel purification=== | ||
| Line 137: | Line 146: | ||
<hr/> | <hr/> | ||
===SLP screening PCR=== | ===SLP screening PCR=== | ||
| - | The PCR run with condition E (5% DMSO, 2mM | + | The PCR run with condition E (5% DMSO, 2mM MgCl<sub>2</sub> with GC buffer) had the highest flexibility; it yields the expected product at different annealing temperatures: 58.4 C, 59.6, C 60.6 C, 61.9 C, 63.2 C, 64.6 C, 65 C. |
Navigate to the [[Team:DTU-Denmark/Notebook/12_August_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/14_August_2013|Next]] Entry | Navigate to the [[Team:DTU-Denmark/Notebook/12_August_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/14_August_2013|Next]] Entry | ||
{{:Team:DTU-Denmark/Templates/EndPage}} | {{:Team:DTU-Denmark/Templates/EndPage}} | ||
Latest revision as of 11:57, 4 October 2013
13 August 2013
Contents |
Lab 208
Main purpose
- PCR to amplify USER fragments of the constitutive SPL;
- PCR of Nir2 with USER primers;
- Gel purification of araBAD SPL.
Who was in the lab
Kristian, Henrike, Gosia
Procedure
PCR (promoter library)
PCR to amplify USER fragments of the constitutive SPL, the constitutive reference promoter and the arabinose reference promoter.
Used 5% DMSO, 2 uL of mM MgCl2 and GC-buffer.
constitutive SPL
- template: pZA21::RFP
- primers: 52a, 52b1
- annealing temp: 58.1C
- elongation time: 3:00
constitutive reference
- template: pZA21::RFP
- primers: 52a, 52b2
- annealing temp: 58.1C
- elongation time: 3:00
Wanted to make negative for the two above reactions but accidentally put the template in the master mix.
arabinose reference
- template: pZA21:araBAD:RFP (labeled Ara)
- primers: 51a,51b1
- annealing temp: 60.3C
- elongation time: 4:00
program (used for all 3 reactions):
| temperature | time | cycles |
|---|---|---|
| 98C | 2:00 | - |
| 98C | 0:20 | 36 |
| annealing temp | 1:00 | 36 |
| 72C | extension time | 36 |
| 72C | 5:00 | - |
| 10C | hold | - |
PCR of Nir2 with USER primers
| sample nr. | buffer | %DMSO | program |
|---|---|---|---|
| 1 | GC | 2 | 55C |
| 2 | GC | 3 | 55C |
| 3 | GC | 5 | 55C |
| 4 | HF | 2 | 55C |
| 5 | HF | 3 | 55C |
| 6 | HF | 5 | 55C |
| 7 | GC | 2 | ramp |
| 8 | GC | 3 | ramp |
| 9 | GC | 5 | ramp |
| 10 | HF | 2 | ramp |
| 11 | HF | 3 | ramp |
| 12 | HF | 5 | ramp |
- PCR mix according to standard protocol with changes: addition of DMSO in 3 different final concentrations (2%, 3%, 5%); two different buffers (HF 5x and GC 5x), amount of added water was dependent on volume of added DMSO.
- Primers for Nir2 - 40a, 40b
- Template - fragment Nir2 amplified with non-uracil primers, 3uL were used
- Polymerase x7
- Program A99, samles 1-6, was based on standard PCR program with 55 C and 1 min of annealing parameters and 5 min of extension time.
- Program A1, samples 7-12, PCR with initial annealing temperature of 59C falling down to 53C (rate 0.1 C/min). 5 min of extension time.
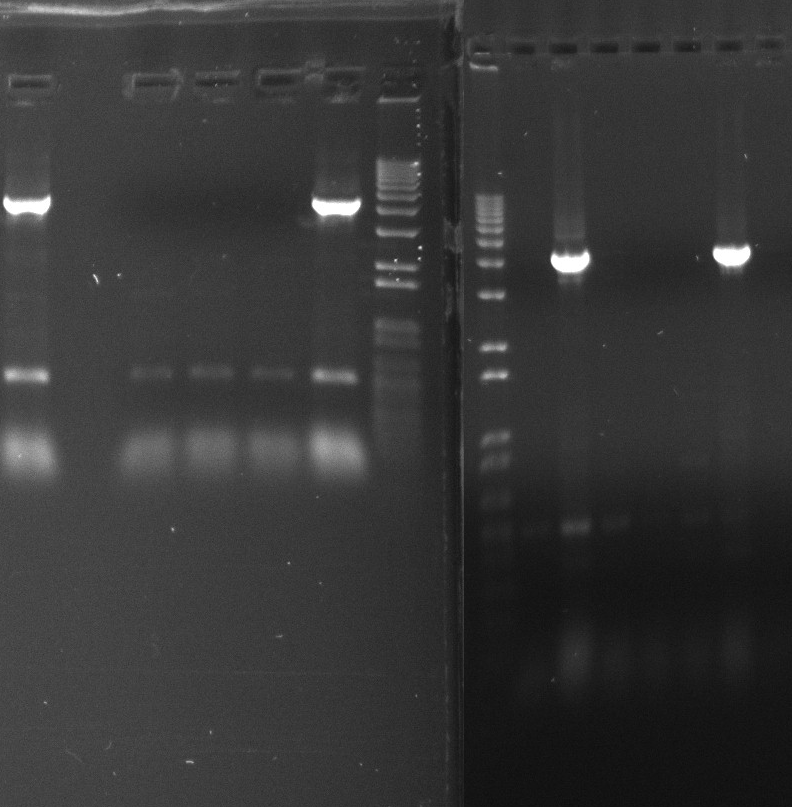
Gel purification
Gel purified araBAD SPL from yesterdays screening PCR
Results
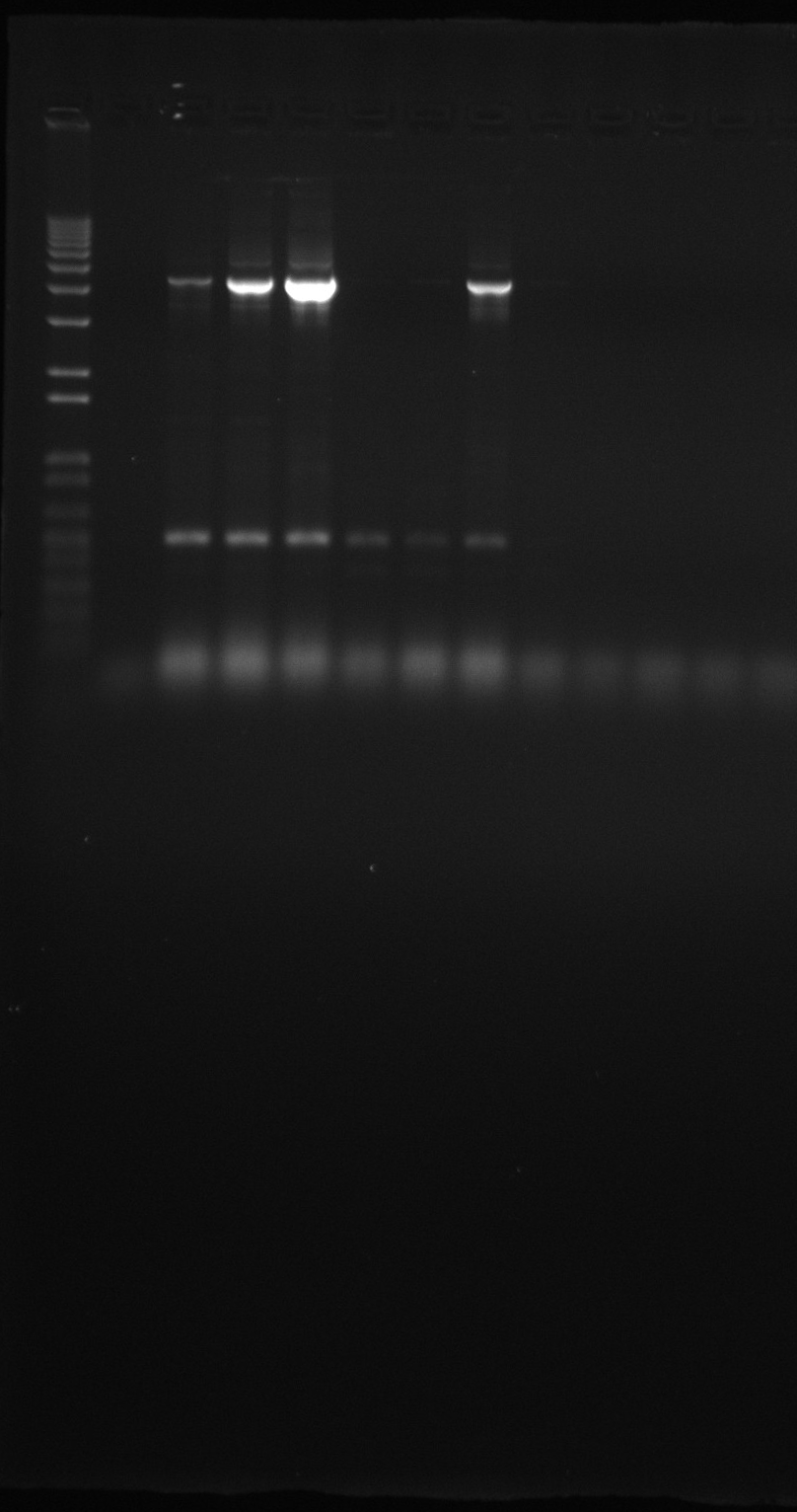
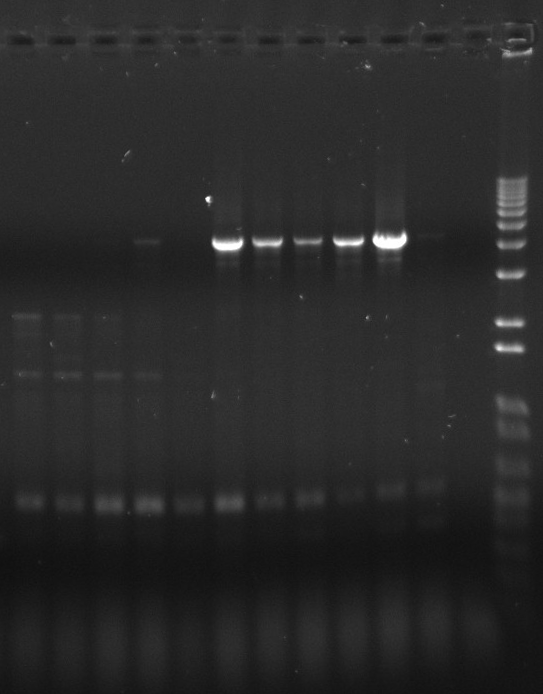
Gel of SLP screening PCR products (yesterday)
Each gel is representing one row; from left to right and up to down is row A to H:
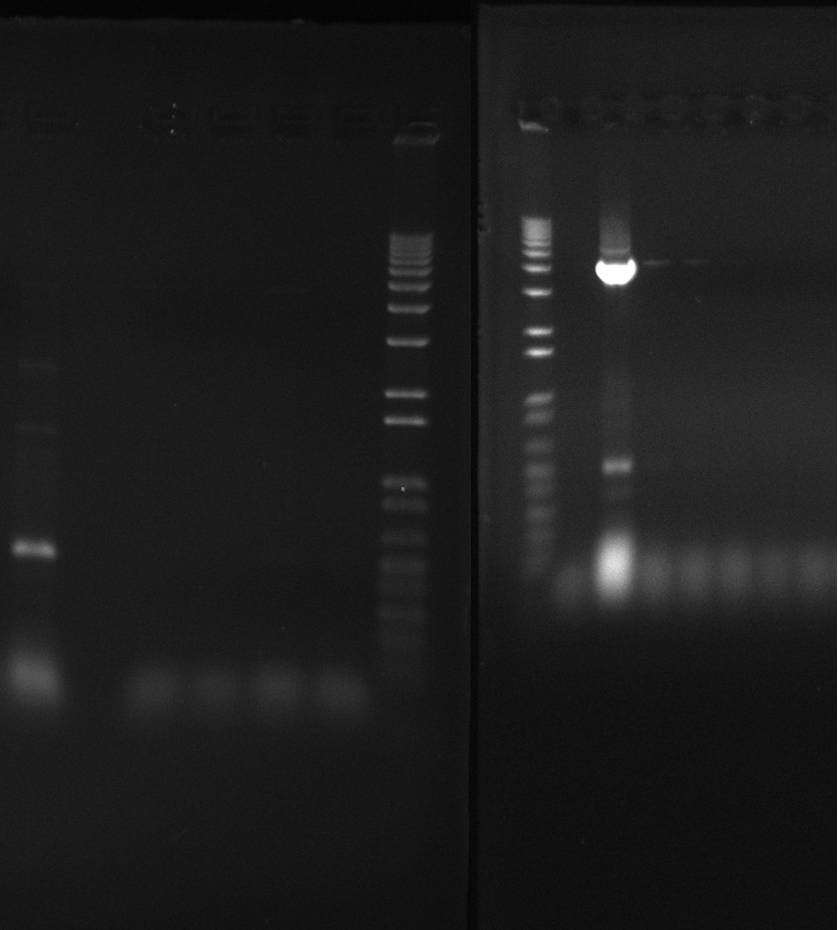
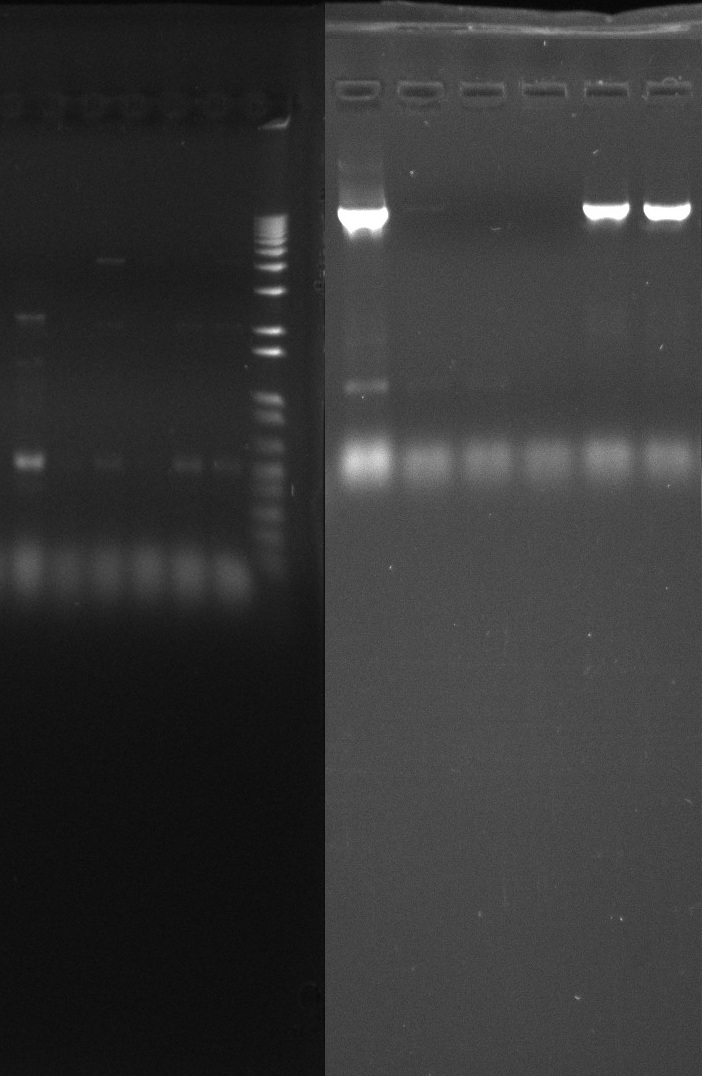
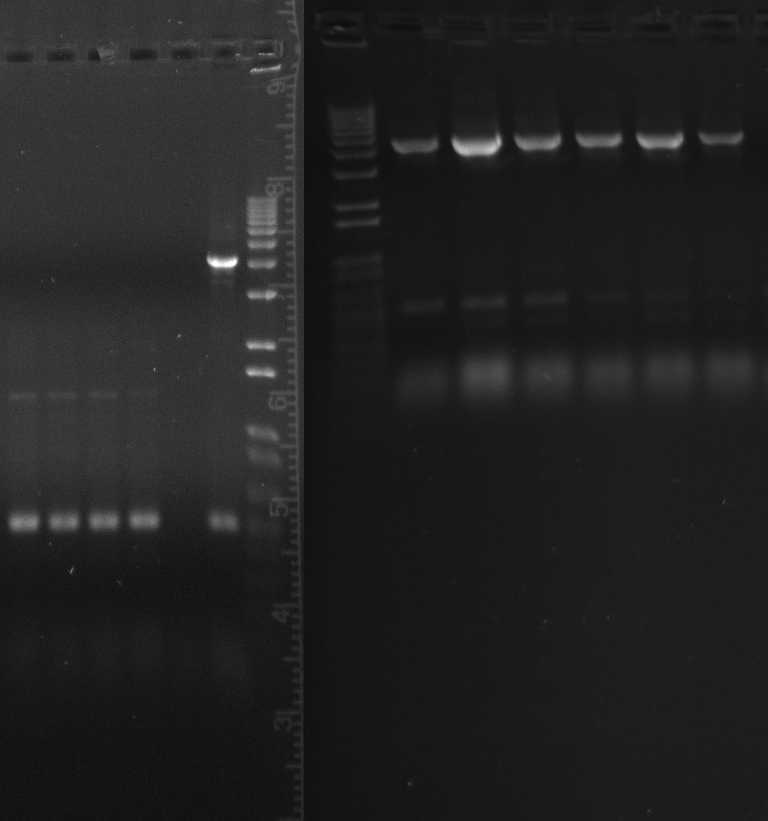
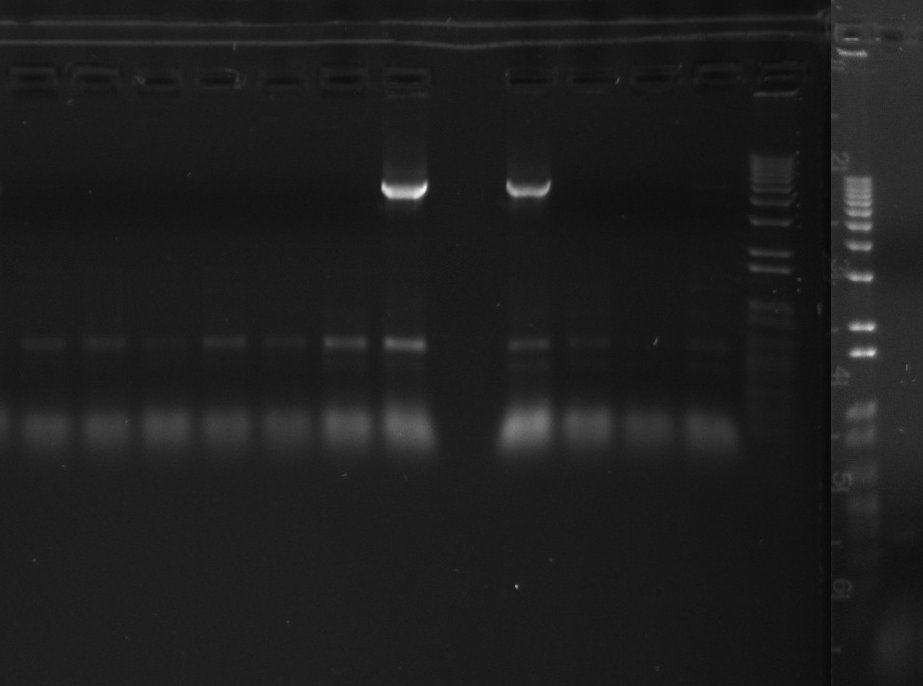
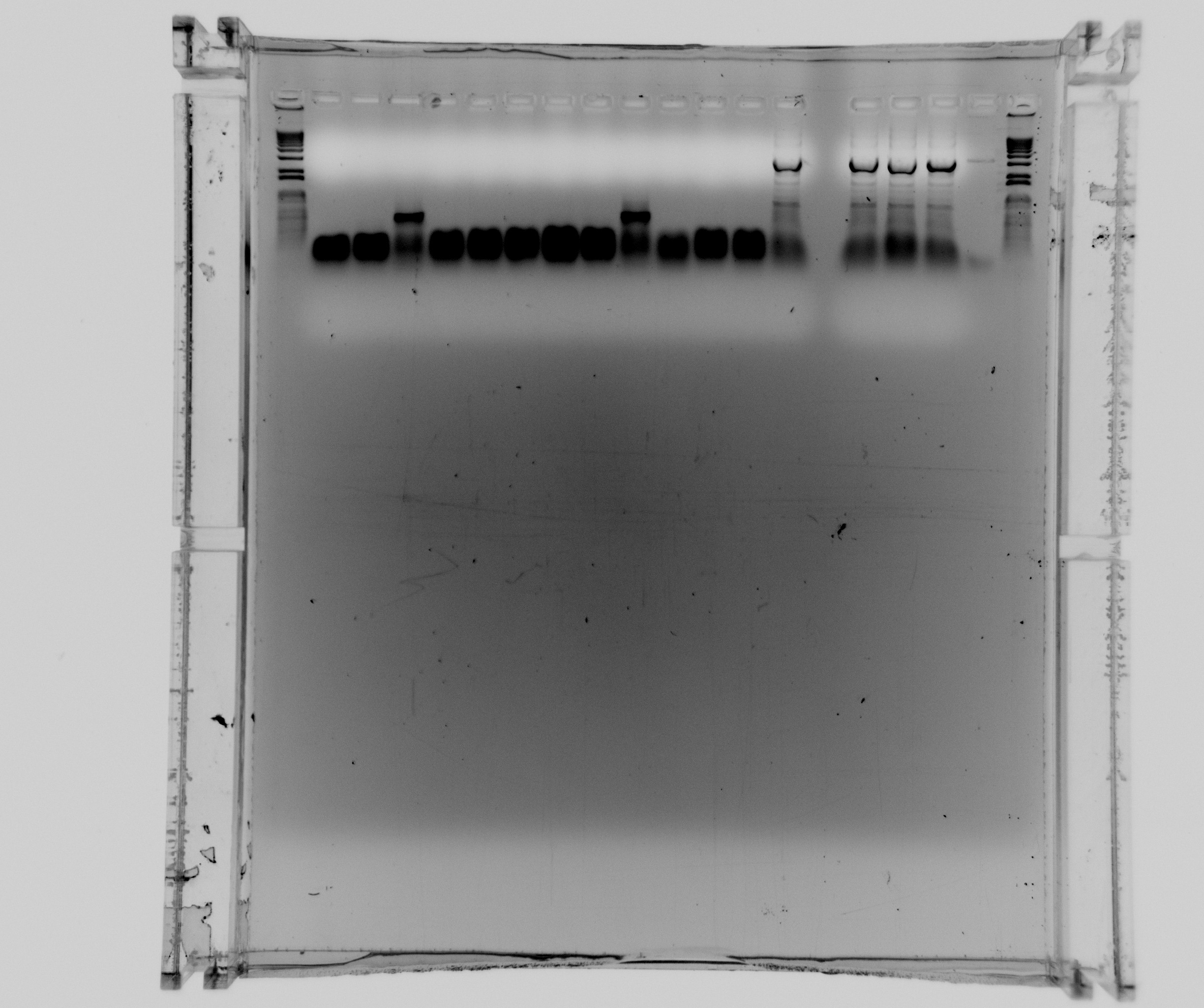
Gel of todays PCR reactions
- 1kb ladder
- Nir2 PCR - sample 1
- Nir2 PCR - sample 2
- Nir2 PCR - sample 3
- Nir2 PCR - sample 4
- Nir2 PCR - sample 5
- Nir2 PCR - sample 6
- Nir2 PCR - sample 7
- Nir2 PCR - sample 8
- Nir2 PCR - sample 9
- Nir2 PCR - sample 10
- Nir2 PCR - sample 11
- Nir2 PCR - sample 12
- constitutive SPL 1
- constitutive SPL 2 (duplicate)
- constitutive promoter reference 1
- constitutive promoter reference 2 (duplicate)
- constitutive promoter reference 3 (very small sample, there was almost nothing left in the master mix)
- 1kb ladder
Conclusion
SLP screening PCR
The PCR run with condition E (5% DMSO, 2mM MgCl2 with GC buffer) had the highest flexibility; it yields the expected product at different annealing temperatures: 58.4 C, 59.6, C 60.6 C, 61.9 C, 63.2 C, 64.6 C, 65 C.
Navigate to the Previous or the Next Entry
 "
"