Team:DTU-Denmark/Notebook/15 August 2013
From 2013.igem.org
(→Results) |
(→Conclusion) |
||
| (10 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:DTU-Denmark/Templates/StartPage|15 August 2013}} | {{:Team:DTU-Denmark/Templates/StartPage|15 August 2013}} | ||
| - | |||
Navigate to the [[Team:DTU-Denmark/Notebook/14_August_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/16_August_2013|Next]] Entry | Navigate to the [[Team:DTU-Denmark/Notebook/14_August_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/16_August_2013|Next]] Entry | ||
| - | + | =Lab 208= | |
| - | = | + | |
<hr/> | <hr/> | ||
| Line 10: | Line 8: | ||
* Plasmid isolation of CycAX, TAT3-2, TAT3-1a, Sec2, HAO, pZA21::RFP and pZA21::araBAD::RFP. | * Plasmid isolation of CycAX, TAT3-2, TAT3-1a, Sec2, HAO, pZA21::RFP and pZA21::araBAD::RFP. | ||
| + | *Nir2 with USER primers | ||
| + | *Prepare araBAD vector for inserts | ||
| + | *Prepare "Hello World" constructs for biobrick integration | ||
| + | |||
==Who was in the lab== | ==Who was in the lab== | ||
| Line 22: | Line 24: | ||
===Glycerol stock cultures=== | ===Glycerol stock cultures=== | ||
-80C stock solution of Sec2, TAT3-1a, TAT3-2 | -80C stock solution of Sec2, TAT3-1a, TAT3-2 | ||
| + | |||
| + | ===DpnI treatment=== | ||
| + | |||
| + | DpnI treated the USER fragments for the SPl project since there were many colonies on the negative controls which can only arise from template DNA used in the PCR. This is removed by DpnI. | ||
| + | |||
| + | ===User ligation and transformation=== | ||
| + | |||
| + | Repeated ligation and transformation from yesterday with same reaction mix to get seperated colonies useable for the SPL project. Plating in a gradient: 5uL, 50 uL, 100uL | ||
| + | |||
| + | ===SPL project=== | ||
| + | |||
| + | Picked single colonies of araBAD SPL transformants and replated them to prepare for Biolector experiment to measure the strength of the expression. | ||
| + | |||
| + | ===Gradient PCR on Nir2=== | ||
| + | Made gradient PCR on Nir2 to get the last USER fragment. The gradient was going from 60C → 72C with 12 reactions. Reactions where all made with same composition; GC-buffer, 2uL 50mM MgCl2 per reaction and 5% DMSO. | ||
| + | |||
| + | ===PCR on pSB1C3=== | ||
| + | Done with primerpair 54 as a two step PCR program with 98C in 10 sec. and 72C for 1:10 min looping 35 times. | ||
| + | |||
| + | ===PCR on constructs for biobrick integration=== | ||
| + | Done with primerpair 53 for all three templates; Sec, TAT3 and cycAX constructs. They where run on same PCR program with 56C annealing for 1 min. and 40 sec. extension. | ||
==Results== | ==Results== | ||
<hr/> | <hr/> | ||
| + | ===gel=== | ||
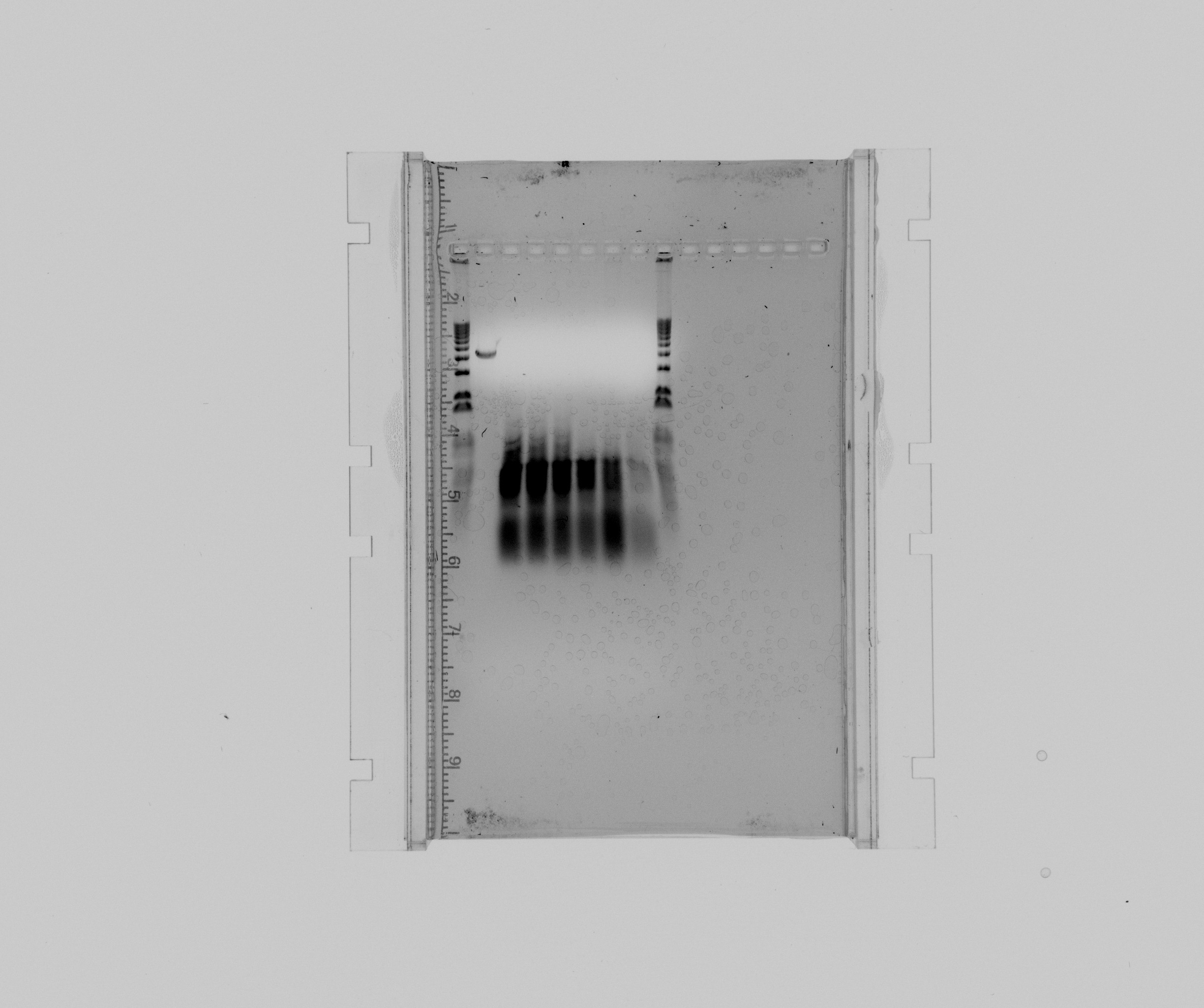
* 1 kb ladder | * 1 kb ladder | ||
* purification of the extraction fragment of Nir2 | * purification of the extraction fragment of Nir2 | ||
| Line 36: | Line 60: | ||
* 1 kb ladder | * 1 kb ladder | ||
| - | == | + | [[File:2013-08-15 nir2 user.jpg|600px]] |
| - | + | ||
| + | The purification has the wrong length for Nir2 and was therefore discarded. | ||
| + | |||
| + | ===transformation=== | ||
| + | |||
| + | Yesterday's transformation yielded too many colonies that were inseparable, it will therefore be repeated and less volume will be plated. | ||
| + | |||
| + | |||
Navigate to the [[Team:DTU-Denmark/Notebook/14_August_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/16_August_2013|Next]] Entry | Navigate to the [[Team:DTU-Denmark/Notebook/14_August_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/16_August_2013|Next]] Entry | ||
{{:Team:DTU-Denmark/Templates/EndPage}} | {{:Team:DTU-Denmark/Templates/EndPage}} | ||
Latest revision as of 11:58, 4 October 2013
15 August 2013
Contents |
Lab 208
Main purpose
- Plasmid isolation of CycAX, TAT3-2, TAT3-1a, Sec2, HAO, pZA21::RFP and pZA21::araBAD::RFP.
- Nir2 with USER primers
- Prepare araBAD vector for inserts
- Prepare "Hello World" constructs for biobrick integration
Who was in the lab
Ariadni, Helen, Kristian, Julia
Procedure
Plasmid isolation
Plasmid have been isolated following the protocol provided by ORIGENE PowerPrep HP Plasmid Miniprep Kit.
Glycerol stock cultures
-80C stock solution of Sec2, TAT3-1a, TAT3-2
DpnI treatment
DpnI treated the USER fragments for the SPl project since there were many colonies on the negative controls which can only arise from template DNA used in the PCR. This is removed by DpnI.
User ligation and transformation
Repeated ligation and transformation from yesterday with same reaction mix to get seperated colonies useable for the SPL project. Plating in a gradient: 5uL, 50 uL, 100uL
SPL project
Picked single colonies of araBAD SPL transformants and replated them to prepare for Biolector experiment to measure the strength of the expression.
Gradient PCR on Nir2
Made gradient PCR on Nir2 to get the last USER fragment. The gradient was going from 60C → 72C with 12 reactions. Reactions where all made with same composition; GC-buffer, 2uL 50mM MgCl2 per reaction and 5% DMSO.
PCR on pSB1C3
Done with primerpair 54 as a two step PCR program with 98C in 10 sec. and 72C for 1:10 min looping 35 times.
PCR on constructs for biobrick integration
Done with primerpair 53 for all three templates; Sec, TAT3 and cycAX constructs. They where run on same PCR program with 56C annealing for 1 min. and 40 sec. extension.
Results
gel
- 1 kb ladder
- purification of the extraction fragment of Nir2
- Nir2 USER touchdown, sample 1
- Nir2 USER touchdown, sample 2
- Nir2 USER touchdown, sample 3
- Nir2 USER touchdown, sample 4
- Nir2 USER touchdown, sample 5
- Nir2 USER touchdown negative
- 1 kb ladder
The purification has the wrong length for Nir2 and was therefore discarded.
transformation
Yesterday's transformation yielded too many colonies that were inseparable, it will therefore be repeated and less volume will be plated.
Navigate to the Previous or the Next Entry
 "
"