Team:DTU-Denmark/Notebook/16 August 2013
From 2013.igem.org
(→Gel on today's PCR for the Biobrick project) |
(→Gradient PCR for Nir2 USER) |
||
| (14 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:DTU-Denmark/Templates/StartPage|16 August 2013}} | {{:Team:DTU-Denmark/Templates/StartPage|16 August 2013}} | ||
| - | + | Navigate to the [[Team:DTU-Denmark/Notebook/15_August_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/19_August_2013|Next]] Entry | |
| - | = | + | =Lab 208= |
<hr/> | <hr/> | ||
==Main purpose== | ==Main purpose== | ||
<hr/> | <hr/> | ||
| + | *Gradient PCR for Nir2 USER | ||
| + | *BioBrick construction | ||
| + | *SPL project | ||
| + | *Preparation for sequencing | ||
==Who was in the lab== | ==Who was in the lab== | ||
| Line 15: | Line 19: | ||
===Gradient PCR for Nir2 USER=== | ===Gradient PCR for Nir2 USER=== | ||
| - | Set up new gradient PCR for Nir2 USER to run in 223. Reaction mix without | + | Set up new gradient PCR for Nir2 USER to run in 223. Reaction mix without MgCl<sub>2</sub> but using DMSO and GC-buffer. |
*template: Nir2 extraction fragment | *template: Nir2 extraction fragment | ||
| Line 22: | Line 26: | ||
*buffer: GC | *buffer: GC | ||
| - | Gradient went from 60C to | + | Gradient went from 60C to 72 <sup>o</sup>C in 12 steps. |
{| class="wikitable" style="text-align: right" | {| class="wikitable" style="text-align: right" | ||
|- | |- | ||
| Line 56: | Line 60: | ||
|- | |- | ||
|} | |} | ||
| + | |||
| + | Note: Redid this PCR with X7 instead of PHUSION polymerase. | ||
<u>pSB1C3</u>: | <u>pSB1C3</u>: | ||
| - | Done with | + | Done with primer pair 54. |
| + | |||
| + | {| class="wikitable" style="text-align: right" | ||
| + | ! temperature !! time !! cycles | ||
| + | |- | ||
| + | | 98C || 2:00 || - | ||
| + | |- | ||
| + | | 98C || 0:10 || 36 | ||
| + | |- | ||
| + | | 58C || 0:20 || 36 | ||
| + | |- | ||
| + | | 72C || 0:35 || 36 | ||
| + | |- | ||
| + | | 72C || 10:00 || - | ||
| + | |- | ||
| + | | 10C || hold || - | ||
| + | |- | ||
| + | |} | ||
===SPL project=== | ===SPL project=== | ||
| - | Picked single colonies of araBAD SPL transformants and replated them to prepare for Biolector experiment to measure the strength of the expression. | + | Picked single colonies of araBAD ref, constitutive SPL and constitutive ref transformants and replated them on plates sprayed with arabinose to prepare for Biolector experiment to measure the strength of the expression. |
===Preparation for sequencing=== | ===Preparation for sequencing=== | ||
| Line 108: | Line 131: | ||
* Nir2 USER, sample 12 | * Nir2 USER, sample 12 | ||
* 1 kb ladder | * 1 kb ladder | ||
| + | |||
| + | [[File:2013-08-16 nir2 user gradient.jpg|600px]] | ||
| + | |||
| + | Samples 5-10 and 12 were cut out for gel purification. | ||
==Conclusion== | ==Conclusion== | ||
<hr/> | <hr/> | ||
| - | Navigate to the [[Team:DTU-Denmark/Notebook/15_August_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/ | + | |
| + | The amplification of the User-ready backbone was sucessfull, but we have to redo the PCR for the signal peptides and the cytochromes. Nir2 with USEr endings was sucessfully amplified. | ||
| + | |||
| + | Navigate to the [[Team:DTU-Denmark/Notebook/15_August_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/19_August_2013|Next]] Entry | ||
{{:Team:DTU-Denmark/Templates/EndPage}} | {{:Team:DTU-Denmark/Templates/EndPage}} | ||
Latest revision as of 11:59, 4 October 2013
16 August 2013
Contents |
Lab 208
Main purpose
- Gradient PCR for Nir2 USER
- BioBrick construction
- SPL project
- Preparation for sequencing
Who was in the lab
Kristian, Julia, Henrike
Procedure
Gradient PCR for Nir2 USER
Set up new gradient PCR for Nir2 USER to run in 223. Reaction mix without MgCl2 but using DMSO and GC-buffer.
- template: Nir2 extraction fragment
- primers: 40a, 40b
- additives: 5% DMSO
- buffer: GC
Gradient went from 60C to 72 oC in 12 steps.
| sample # | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| temperature | 60.2 | 60.5 | 61.6 | 63.0 | 64.2 | 65.4 | 66.6 | 67.6 | 68.9 | 70.2 | 71.6 | 72 |
PCR for BioBrick construction
PCR amplified the constructs from Hello World and cytochromes and pSB1C3, which will be ligated together into biobricks.
constructs:
- GC-buffer, 5%DMSO, 2uL 50mM MgCl2
- primers: 53a, 53b
- templates: TAT3-2, Sec2, cycAX
Tried two different annealing temperatures. Program:
| temperature | time | cycles |
|---|---|---|
| 98C | 2:00 | - |
| 98C | 0:10 | 36 |
| 60C/56C | 0:45 | 36 |
| 72C | 0:45 | 36 |
| 72C | 5:00 | - |
| 10C | hold | - |
Note: Redid this PCR with X7 instead of PHUSION polymerase.
pSB1C3:
Done with primer pair 54.
| temperature | time | cycles |
|---|---|---|
| 98C | 2:00 | - |
| 98C | 0:10 | 36 |
| 58C | 0:20 | 36 |
| 72C | 0:35 | 36 |
| 72C | 10:00 | - |
| 10C | hold | - |
SPL project
Picked single colonies of araBAD ref, constitutive SPL and constitutive ref transformants and replated them on plates sprayed with arabinose to prepare for Biolector experiment to measure the strength of the expression.
Preparation for sequencing
Diluted miniprepped plasmids down to 100 ng/uL. Have to redo cycAx in pZA21 purification because it was diluted too much.
Results
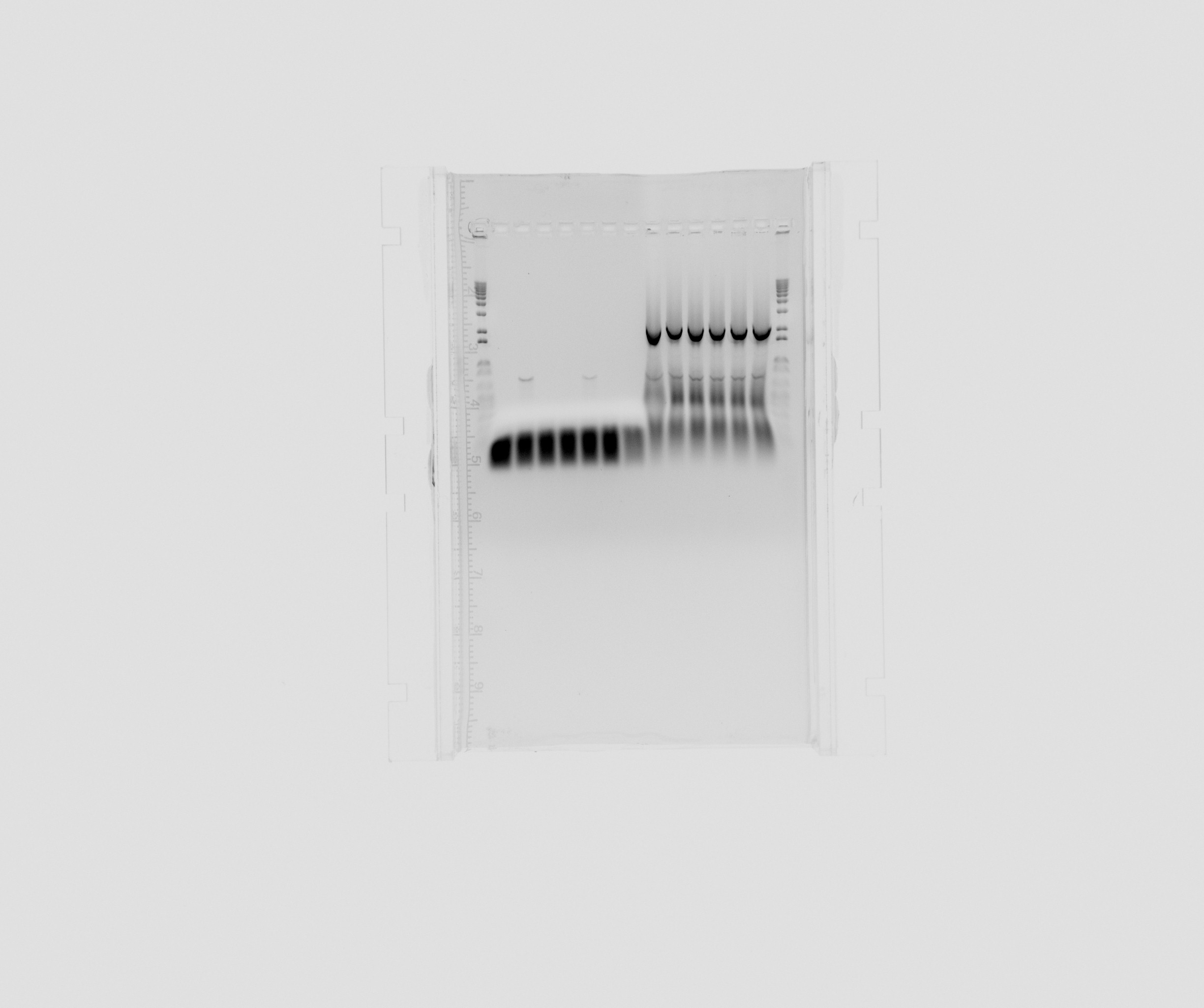
Gel on today's PCR for the Biobrick project
- 1 kb ladder
- Sec2 on 60C
- TAT3-2 on 60C
- cycAX on 60C
- Sec2 on 56C
- TAT3-2 on 56C
- cycAX on 56C
- negative for construct parts
- pSB1C3 backbone
- pSB1C3 backbone
- pSB1C3 backbone
- pSB1C3 backbone
- pSB1C3 backbone
- pSB1C3 backbone
- 1 kb ladder
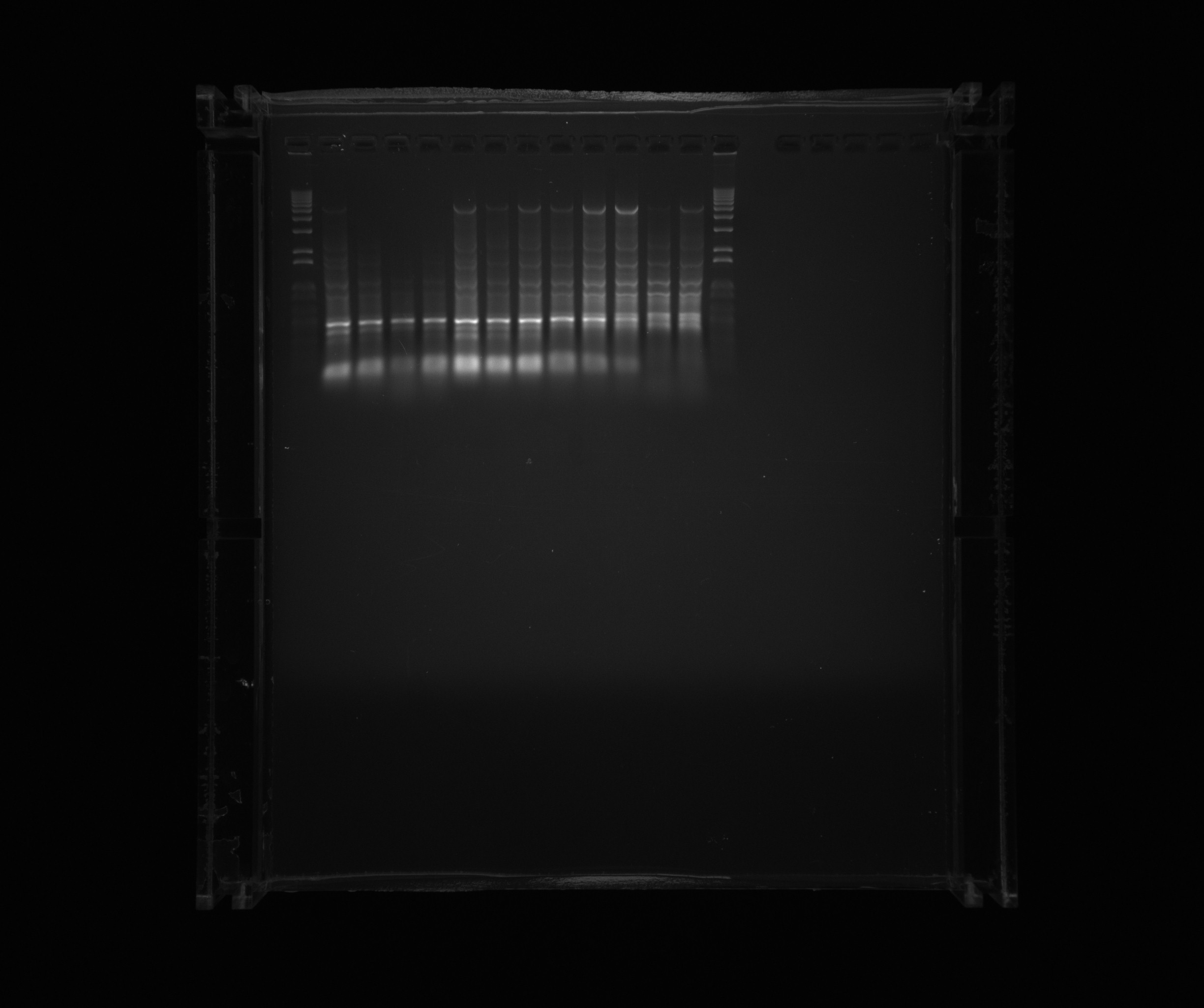
Gel on screening PCR to amplify Nir2 USER
- 1 kb ladder
- Nir2 USER, sample 1
- Nir2 USER, sample 2
- Nir2 USER, sample 3
- Nir2 USER, sample 4
- Nir2 USER, sample 5
- Nir2 USER, sample 6
- Nir2 USER, sample 7
- Nir2 USER, sample 8
- Nir2 USER, sample 9
- Nir2 USER, sample 10
- Nir2 USER, sample 11
- Nir2 USER, sample 12
- 1 kb ladder
Samples 5-10 and 12 were cut out for gel purification.
Conclusion
The amplification of the User-ready backbone was sucessfull, but we have to redo the PCR for the signal peptides and the cytochromes. Nir2 with USEr endings was sucessfully amplified.
Navigate to the Previous or the Next Entry
 "
"