Results
Over the summer Team Magneto worked hard and some of the preset goals were successfully accomplished. Here we show and explain you our main results.
Quick Overview
MamC-eGFP: the functionality of the eGFP part of the fusion was proven, regarding the MamC part only the expected size of the protein could be shown
MamC alone couldn’t be further characterized
eGFP: the functionality and autenticity of the protein could be shown
eFbFP: the submitted BioBrick couldn’t be successfully characterized
MamC-eGFP
MamC (BBa_K1094001) from Magnetospirillum magnetotacticum (MS-1) was tagged with enhanced green fluorescent protein (eGFP, BBa_K1094400). Between the parts a glycine linker (10 glycine residues) is added. The gene product can be used to detect localization of MamC in MS-1. The two parts were cloned together using classical cloning and the composite part was subsequently cloned into pJAM1786 using the Gateway system. Colony PCR and gel electrophoresis was performed to ensure the insert was in the vector (Fig.1).
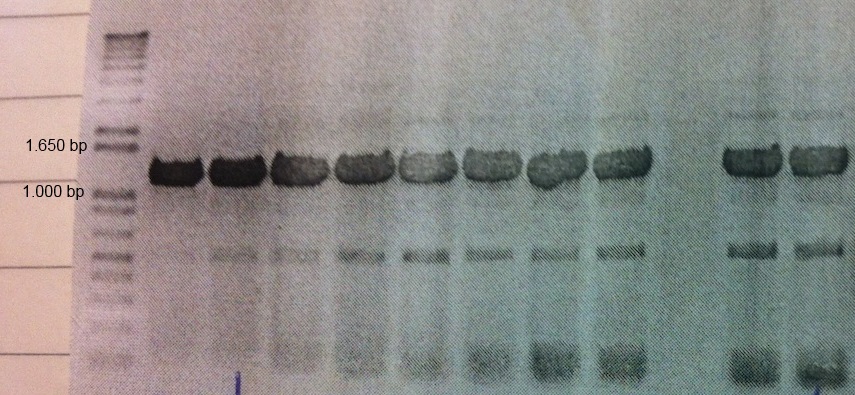
Figure 1: Colony PCR and gel electrophoresis was performed to ensure the insert was in the vector. The figure shows clear bands at around the expected size of 1.2 kb
In order to demonstrate that the fusion protein was both expressed and functional, we made an SDS page followed by western blot analysis and we also observed a fluorescence signal by using confocal microscopy. Moreover, fluorescence measurements were made on the ELISA reader.

Figure 2: Western Blot analysis of transformed E.coli cultures with anti-GFP antibody. Shows expected band of MamC-eGFP at around 45 kDa and a band for cleaved off eGFP at around 32 kDa.
The western blot analysis (Fig.2) shows the presence of two bands, one at around 45 kDa and the other one at approximately 32 kDa. These results confirm the expected molecular weight for MamC-eGFP which is 45 kDa. The second band would be eGFP, because it has the molecular weight of 32,7 kDa and it probably derives from the cleavage of the Gly-linker used between MamC and eGFP.

Figure 3: Left fluorescence signal from MamC- eGFP observed through confocal microscopy. On the right a white field view onto the same E.coli cells.
Looking at the results of the confocal microscopy, we can clearly observe fluorescence corresponding with the E.coli cells. This image (Fig. 3) attests the functionality of the eGFP part of the fusion protein.
For the fluorescence measurements eight transformed colonies were inoculated into liquid culture and grown overnight alongside three E. coli cultures containing a non-expression plasmid with a non-fluorescent insert (MamC-pSB1C3). Fluorescence was measured on an ELISA reader.
The mean values of the fluorescence can be found in the table (Tab. 1) below. Standard error for both groups (Total SE) was assessed using a Satterthwaite approximation.
Table 1: Showing fluorescence measurements on MamC-eGFP.

All dilution showed significantly more fluorescence than the control cultures except for the 50X dilution (Fig. 4). The mean of the difference to the control cultures is shown as a function of dilution below. These results additionally support the functionality of our construct.

Figure 4: Plot of the fluorescence of MamC-eGFP in relation to a negative control.
The fluorescence measurements and confocal images support the conclusion that the eGFP part of the MamC-eGFP fusion is functional, along with the western blot which also proves the right size of the MamC portion of the construct.
MamC
The MamC sequence was taken from Magnetospirillum magnetotacticum’s (MS-1) genomic DNA. The protein is supposed to be located in the magnetosome membrane thus making MamC ideal for anchoring foreign proteins to the magnetosome.
There were no results obtained on the MamC protein alone, since we didn’t have any possibility to prove its functionality.
Enhanced Green Fluorescent Protein (eGFP)
Enhanced GFP is a modified version of the green fluorescent protein (GFP) derived from Aequorea victoria with enhanced fluorescence properties. These enhanced fluorescent properties of the eGFP dramatically improved the spectral characteristics of GFP, resulting in increased fluorescence.
The part was cloned into the expression vector pJAM1786 in E. coli, by using the Gateway system. Colony PCR and gel electrophoresis was performed to ensure the insert was in the vector (shown in the seven leftmost lanes of the figure 5 below).
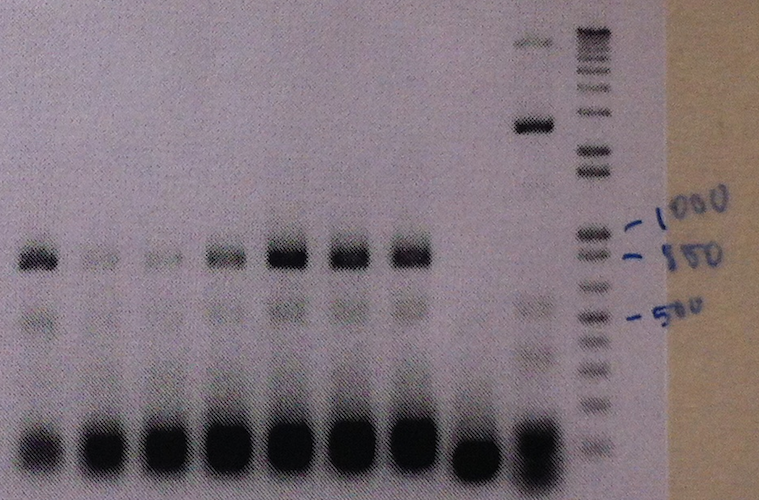
Figure 5: Colony PCR and gel electrophoresis was performed to ensure the insert was in the vector. The figure shows clear bands at around the expected size of 800 bp.
The restreaks of those seven colonies were inoculated into liquid culture and grown overnight alongside three E. coli cultures containing an empty plasmid (pBBR1MCS-2). Fluorescence was measured in an opaque ELISA reader plate.
The mean values of the fluorescence can be found in the table (Tab.2) below. Standard error for both groups (Total SE) was assessed using a Satterthwaite approximation.
Table 2: Showing fluorescence measurements on eGFP.

All dilution showed significantly more fluorescence than the control cultures except for the 50X dilution. The mean of the difference to the control cultures is shown as a function of dilution below (Fig.6).

Figure 6: Plot of the fluorescence of eGFP in relation to a negative control.
To further show the expression and functionality of our eGFP a western blot with an anti-GFP antibody was carried out (Fig. 7).

Figure 7: Western Blot analysis of transformed E.coli cultures with anti-GFP antibody. Shows expected band of eGFP next to the positive control sample (GFP)
The western blot shows the expected band for eGFP at around 32,7 kDa and next to the GFP sample that was used as positive control. Thereby supporting the authenticity of the protein.
As a next step E.coli cultures carrying the eGFP construct were observed under the confocal microscope for fluorescence (Fig.8).

Figure 8: Left fluorescence signal from eGFP observed through confocal microscopy. On the right a white field view onto the same E. coli cells.
The images made by confocal microscopy prove the functionality of the eGFP along with the results from the fluorescence measurements and the western blot.
Enhanced flavin-binding fluorescent protein (eFbFP)
The reporter eFbFP is a fluorescent protein that uses flavin mono nucleotide (FMN) as a cofactor. The reporter is suitable for use in anaerobic conditions.
The part was cloned into expression vector pJAM1786 using the Gateway system. The plasmid was transformed into E. coli cells. Colony PCR was carried out and colonies were inoculated into liquid culture. The following day the colonies failed to show significantly more fluorescence than three control cultures.
This was unexpected since we previously showed significant fluorescence from a very similar sequence. Due to two internal PstI sites that sequence was not BioBrick compatible. After removing the restriction sites fluorescence was lost even though no change in amino acid sequence was made. Therefore, we expect the lack of fluorescence to be caused by cloning/transformation/technical issues and not the sequence. Due to time limitation we were not able to support this statement.
Conclusion
The eGFP protein was fully characterized. In fusion with eGFP proof of the correct size of the MamC product could be presented. MamC on its own could unfortunately not be characterized due to technical limitations. eFbFP lost its functionality after the removal of internal restriction sites and therefore its functionality could not be shown here.
Outlook
For presenting improved results in November at MIT in Boston, we would work on characterizing MamC by acquiring a MamC antibody, which is already commercially available. As a next step the MamC-eGFP construct would need to be conjugated into magnetotactic bacteria to show that it localizes as expected to the magnetosome membrane. Also the eFbFP construct would get debugged and also characterized via fluorescence measurements and western blot analysis for which we would need to develop an antibody or clone eFbFP with an incorporated tag (myc, flag or his).
 "
"


