Team:Heidelberg/Tyrocidine week15 ms
From 2013.igem.org
(Created page with " ==DNA concentrations of fragments== ===Analysis of DNA concentration=== right * Concentrations obtained through quantifica...") |
|||
| Line 1: | Line 1: | ||
| + | |||
==DNA concentrations of fragments== | ==DNA concentrations of fragments== | ||
| Line 4: | Line 5: | ||
===Analysis of DNA concentration=== | ===Analysis of DNA concentration=== | ||
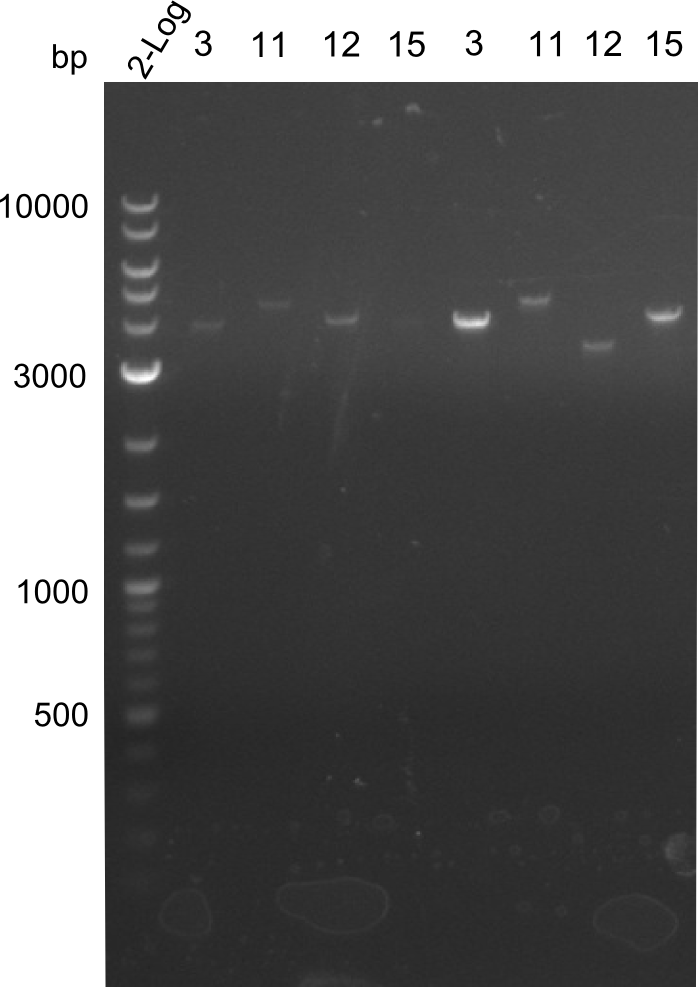
| - | [[File: | + | [[File:Heidelberg_Tyr quantification 3 11 12 13 15.png|150px|right]] |
* Concentrations obtained through quantification gel. There are 20µl of all fragments. | * Concentrations obtained through quantification gel. There are 20µl of all fragments. | ||
| Line 152: | Line 153: | ||
===Pictures of plates=== | ===Pictures of plates=== | ||
| - | [[File: | + | [[File:Heidelberg_Plate 12 Tyc.JPG|center|thumbnail|picture of colonies taken on 06.08.2013]] |
===missing: Tetrapeptide I=== | ===missing: Tetrapeptide I=== | ||
| Line 178: | Line 179: | ||
==Validation of Gibson Assembly== | ==Validation of Gibson Assembly== | ||
===Restriction digest of Tripetide I, II and Tetrapeptide II=== | ===Restriction digest of Tripetide I, II and Tetrapeptide II=== | ||
| - | [[File: | + | [[File:Heidelberg_Tyr restriction digest.png|100px|thumb|right|Samples were digested with 0.5 µl NotI. Lanes 2, 8, 10, 11, 20, 25, 37, 40, 43 seem to be positive and are used for futher analysis by restriction with other enzymes]] |
* Minipreps of colonies picked on 06.08.2013 | * Minipreps of colonies picked on 06.08.2013 | ||
| Line 204: | Line 205: | ||
===Restriction Digest of Tri- Di and Tetrapeptides=== | ===Restriction Digest of Tri- Di and Tetrapeptides=== | ||
| - | [[File: | + | [[File:Heidelberg_Restriction disgest SphI MfeI.png|100px|thumb|right|Samples were digested with 1 µl SphI (2 years over due date) (for tri- and tetrapeptides) and MfeI (for dipeptide. All lanes show unexpected lines, however, adding the fragment-sizes leads to expected plasmid size for tetrapeptide]] |
Restriction with MfeI and NotI was performed with samples from miniprep on 07.08.2013 that appeared positive: | Restriction with MfeI and NotI was performed with samples from miniprep on 07.08.2013 that appeared positive: | ||
| Line 246: | Line 247: | ||
=== Proving the results of Gibson Assembly === | === Proving the results of Gibson Assembly === | ||
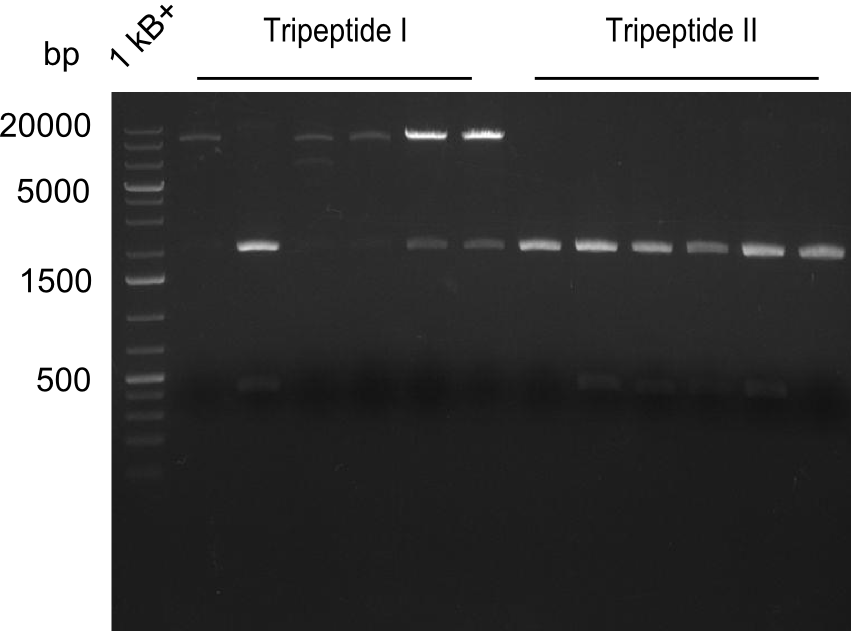
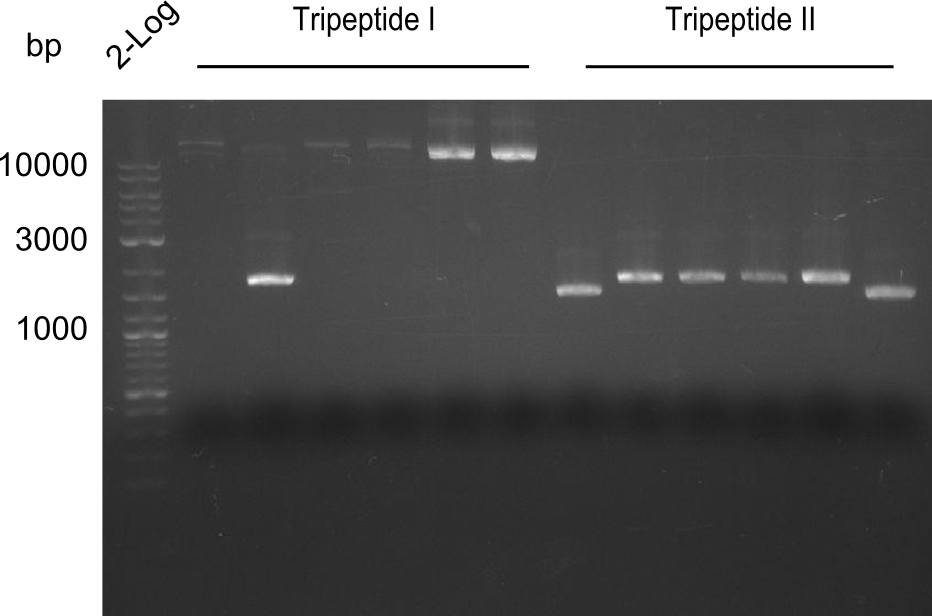
| - | [[File: | + | [[File:Heidelberg_20130809 restriction NotI TriI TriII.png|100px|thumb|right|restriction with NotI]] |
| - | [[File: | + | [[File:Heidelberg_20130809 restriction SphI TriI TriII.png|100px|thumb|right|restriction with SphI]] |
* Colonies from Gibson-Assembly were picked and grown in 3 ml 2x YT medium | * Colonies from Gibson-Assembly were picked and grown in 3 ml 2x YT medium | ||
* Minipreps from Tripeptide-Colonies picked the day before that were grown in 2x YT medium | * Minipreps from Tripeptide-Colonies picked the day before that were grown in 2x YT medium | ||
| Line 282: | Line 283: | ||
* --> see [[Media:Overview Gibson colonies.xlsx|the Gibson construct overview table]] for reference | * --> see [[Media:Overview Gibson colonies.xlsx|the Gibson construct overview table]] for reference | ||
| - | [[File: | + | [[File:Heidelberg_20130810 Restriction Digest Tetrapeptide I Colony.png|100px|thumb|right|restriction with Pst1 and EcoR1]] |
| - | [[File: | + | [[File:Heidelberg_20130810 second Restriction Digest Tetrapeptide I Colony.png|100px|thumb|right|restriction with MfeI]] |
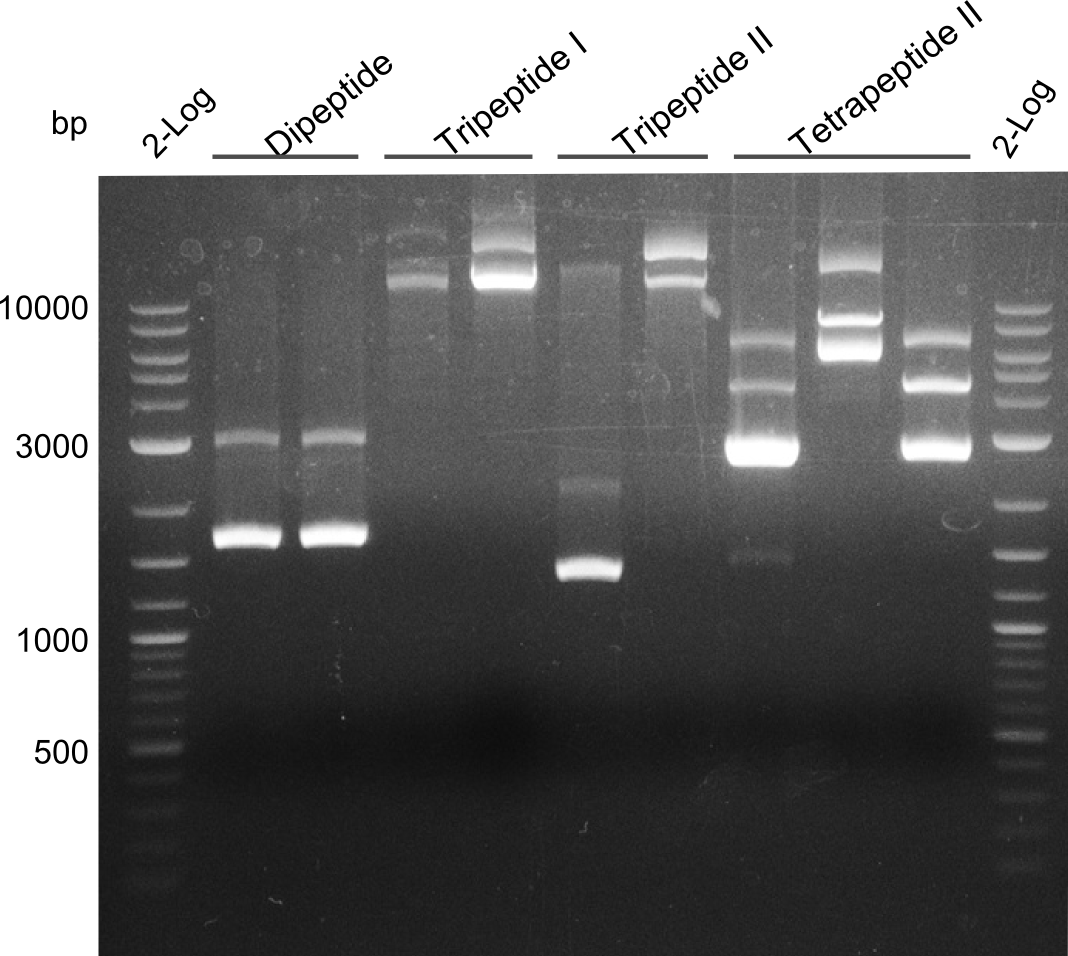
| - | [[File: | + | [[File:Heidelberg_Quantification gel minipreps TetI Di TriI TetII.jpg|100px|thumb|right|quantification gel of minipreps of different short-peptide-NRPSs]] |
{| class="wikitable" | {| class="wikitable" | ||
Latest revision as of 16:20, 4 October 2013
Contents |
DNA concentrations of fragments
Analysis of DNA concentration
- Concentrations obtained through quantification gel. There are 20µl of all fragments.
| fragment | concentration [ng/µl] |
|---|---|
| 3 | 2 |
| 11 | 3 |
| 13 | 7 |
| 15 | -- |
| 3 | 35 |
| 11 | 10 |
| 12 | 10 |
| 15 | 20 |
Complete List of DNA concentrations of all fragments
| fragment | concentration [ng/µl] |
|---|---|
| 1 | 10 (25.7.), 30 (28.7.) |
| 2 | 16 (25.7.), 12 (31.7.) |
| 3 | 35 (04.8.) |
| 4 | 32 (24.7.), 30 (28.7.) |
| 5 | 12 (26.7.), 14 (31.7.) |
| 6 | 8 (26.7.) (empty), 6.5 (03.8.) |
| 7 | 12 (27.7.), 18 (31.7.) |
| 8 | 8 (26.7.), 5 (03.8.) |
| 9 | 30 (28.7.) |
| 10 | 40 (28.7.) |
| 11 | 5 (31.7.), 10 (04.8.) |
| 12 | 10 (04.8.) |
| 13 | 20 (31.7.), 7 (03.8.) |
| 14 | 40 (28.7.) |
| 15 | 11 (31.7.), 20 (04.8.) |
Gibson Assembly
| fragments (10µL) + 10 µL Gibson Mix |
fragment date length
- 29.7 2500
- 29.7 2000
- 5.8. 4000
- 29.7 2500
- 29.7 3000
- 29.7 7000
- 31.7 2000
- 29.7 7000
- 29.7 2500
- 29.7 2000
- 5.8. 5000
- 5.8. 3000
- 31.7 4000
- 29.7 2000
- 5.8. 4000
Dipeptide
| fragment | concentration [ng/µl] | volume for gibson assembly [µL] |
|---|---|---|
| 1 | 30 | 3.14 |
| 2 | 16 | 4.71 |
| 3 | 35 | 2.15 |
Tripeptide I
| fragment | concentration [ng/µl] | volume for gibson assembly [µL] |
|---|---|---|
| 4 | 32 | 1.02 |
| 5 | 12 | 3.27 |
| 6 | 8 | 5.71 |
Tripeptide II
| fragment | concentration [ng/µl] | volume for gibson assembly [µL] |
|---|---|---|
| 4 | 32 | 1.25 |
| 7 | 18 | 1.77 |
| 8 | 8 | 6.98 |
Tetrapeptide II
| fragment | concentration [ng/µl] | volume for gibson assembly [µL] |
|---|---|---|
| 9 | 30 | 1.32 |
| 10 | 40 | 0.79 |
| 11 | 10 | 3.95 |
| 14 | 10 | 0.79 |
| 15 | 20 | 3.16 |
Pictures of plates
missing: Tetrapeptide I
| fragment | concentration [ng/µl] | volume for gibson assembly [µL] |
|---|---|---|
| 9 | 30 | 0.77 |
| 10 | 40 | 0.46 |
| 11 | 10 | 2.31 |
| 12 | 10 | 2.77 |
| 13 | 20 | 3.69 |
overview over Gibson constructs
Excel-Document with constructs, names, DNA-concentrations and important indications
Validation of Gibson Assembly
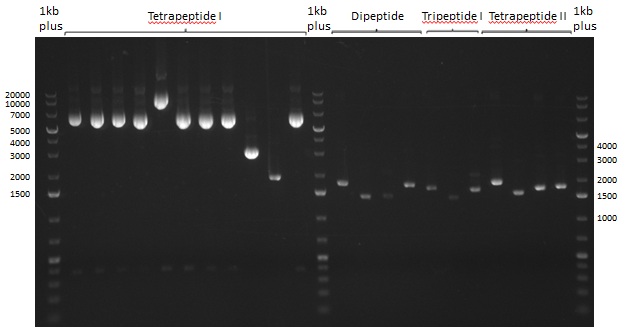
Restriction digest of Tripetide I, II and Tetrapeptide II
- Minipreps of colonies picked on 06.08.2013
- Restriction digest of DNA obtained by minipreps
- with NotI
- reamplifications of fragments 6 and 8 according to Protocol-page
- did not lead to any usable DNA-sample
| what | µl |
|---|---|
| DNA | 4 (~200-800ng) |
| Cutsmart Mix | 3 |
| Enzyme | 1 |
| ddH20 | ad 30 µl |
Results
Gel-Picture taken on 07.08.2013. Different peptide constructs are annotated. For exact reference to picked colonies, see: our overview over our Gibson colonies
Restriction Digest of Tri- Di and Tetrapeptides
Restriction with MfeI and NotI was performed with samples from miniprep on 07.08.2013 that appeared positive:
| what | µl |
|---|---|
| DNA | 1200ng |
| Cutsmart Mix | 2 |
| Enzyme | 1 |
| ddH20 | ad 20 µl |
Gibson Assembly was performed with fragments needed for Tetrapeptide I according to the protocol described above.
| fragment | concentration [ng/µl] | volume for gibson assembly [µL] |
|---|---|---|
| 9 | 30 | 0.77 |
| 10 | 40 | 0.46 |
| 11 | 10 | 2.31 |
| 12 | 10 | 2.77 |
| 13 | 20 | 3.69 |
Results
As the observed bands were not expected, other colonies (Tripeptide I and II) were picked for further analysis. Samples will be send to sequencing (which????xxxxxxxxx).
Proving the results of Gibson Assembly
- Colonies from Gibson-Assembly were picked and grown in 3 ml 2x YT medium
- Minipreps from Tripeptide-Colonies picked the day before that were grown in 2x YT medium
- Restriction digests:
- NotI (~0.8µl per sample)
- SphI (~0.5µl per sample) (enzyme 2 years over due-date)
Results
- ==> Tripeptide I shows a good band for the insert and the backbone, Tripeptide II doesn't show any positive bands
- ==> the SphI-digest was not successful - maybe because enzyme was not working
- for reference, see the overview Excel-document
- samples were sent to sequencing:
- Dipeptide 1 (lane 2) --> from plate Dipeptide 1 (colony B) (picked on 05.08.)
- Dipeptide 2 (lane 8) --> from plate Dipeptide 3 (colony B) (picked on 05.08.)
- Tripeptide I 1 (lane 10) --> from plate Tripeptide I 1 (colony A) (picked on 05.08.)
- Tripeptide I 2 (lane 11) --> from plate Tripeptide I 1 (colony B) (picked on 05.08.)
- Tripeptide II 1 (lane 20) --> from plate Tripeptide II 1 (colony B) (picked on 05.08.)
- Tripeptide II 2 (lane 25) --> from plate Tripeptide II 3 (colony A) (picked on 05.08.)
- Tetrapeptide II 1 (lane 37) --> from plate Tetrapeptide II' 1 (colony A) (picked on 05.08.)
- Tetrapeptide II 2 (lane 40) --> from plate Tetrapeptide II' 2 (colony A) (picked on 05.08.)
- Tripeptide I new 1 (lane 5 on gel image 09.08.2013) --> from plate Tripeptide I 3 (colony C) (picked on 08.08.)
- Tripeptide I new 2 (lane 6 on gel image 09.08.2013) --> from plate Tripeptide I 3 (colony D) (picked on 08.08.)
Glycerol Stock
Glycerol Stocks of all colonies that were sent to sequencing
Glycerol Stocks of newest Di-, Tri- and Tetrapeptide - colonies ->the Gibson construct overview table for reference
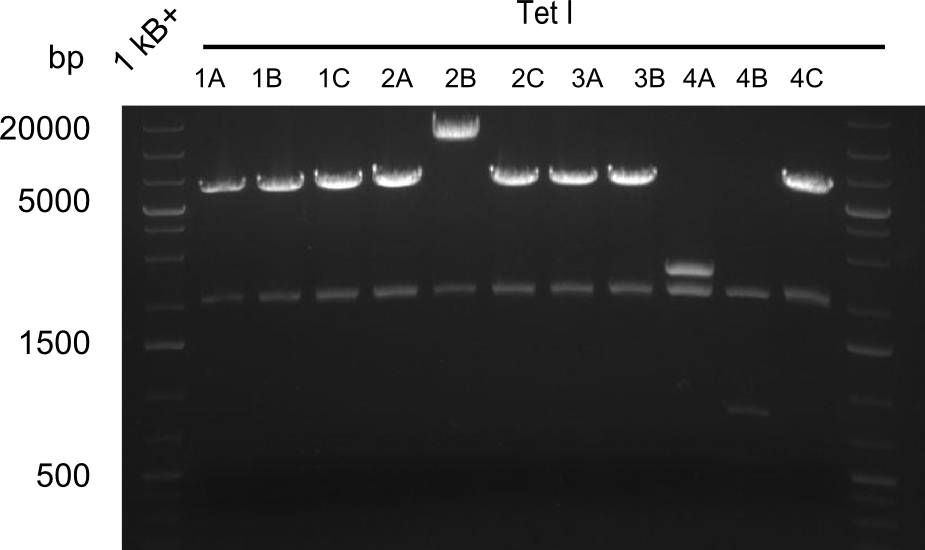
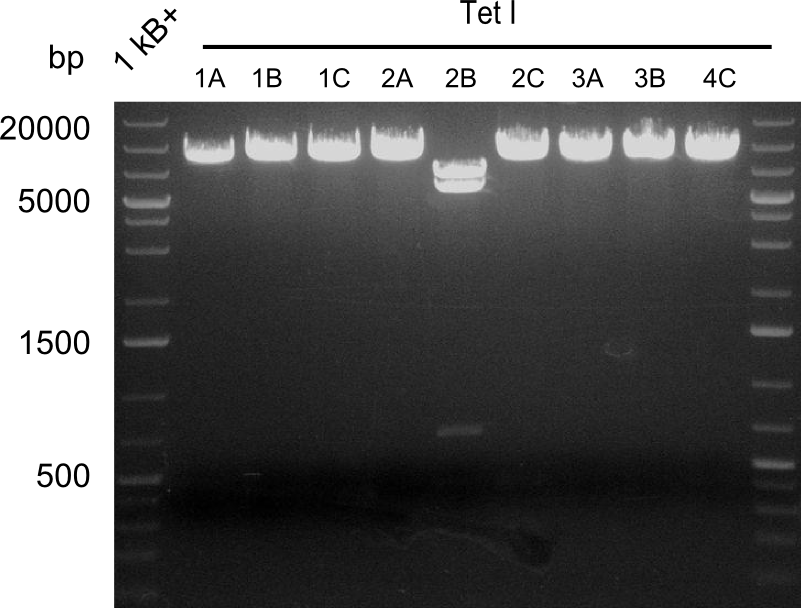
Restriction Digest of Tetrapeptide-I-NRPSs
- Minipreps of Tetrapeptide I colonies and new Di-, Tri- & Tetrapeptide colonies
- --> see the Gibson construct overview table for reference
| Probe name | Available volume |
|---|---|
| Tet I 1a | 1,3 ml |
| Tet I 1b | 1,3 ml |
| Tet I 1c | 1,3 ml |
| Tet I 2a | 1,3 ml |
| Tet I 2b | 1,3 ml |
| Tet I 2c | 1,3 ml |
| Tet I 3a | 0,845 ml |
| Tet I 3b | 0,975 ml |
| Tet I 4a | 1,3 ml |
| Tet I 4b | 0,75 ml |
| Tet I 4c | 0,845 ml |
| Tet II 2 11 | 0,750 ml |
| Tet II 2 14 | 0,975 ml |
| Tet II 3 15 | 0,750 ml |
| Tet II 2 12 | 0,975 ml |
| Di 1 | 0,975 ml |
| Tri II 1 | 0,975 ml |
| Di 2 | 1,075 ml |
| Tri II 2 | 0,975 ml |
| Di 3A | 0,975 ml |
| Tri II 3 | 0,975 ml |
| Di 3b | 0,975 ml |
Results
Restriction digest I of Tetrapeptide I with PstI and EcoR1-HF was probably positive for probe 2b,so we drove another Restriction digest with MfeI for Validation. The result was not clear, perhaps because of the extremely high DNA concentration of our Miniprep or because of the old enzyme.
We decided to sent this probe to sequencing to be sure.
Second Restriction digest with MFE 1 for validation of 2B.
 "
"