Team:Imperial College/mainresults
From 2013.igem.org
Margarita K (Talk | contribs) |
|||
| Line 1: | Line 1: | ||
{{:Team:Imperial_College/Templates:header}} | {{:Team:Imperial_College/Templates:header}} | ||
| + | __NOTOC__ | ||
| + | <h1>Main Results</h1> | ||
| - | < | + | <html> |
| + | <div id="TabbedPanels1" class="TabbedPanels"> | ||
| + | <ul class="TabbedPanelsTabGroup"> | ||
| + | <li class="TabbedPanelsTab" tabindex="0"><h4>Resource-full Waste</h4></li> | ||
| + | <li class="TabbedPanelsTab" tabindex="0"><h4>Plastic Fantastic</h4></li> | ||
| + | |||
| + | </ul> | ||
| + | <div class="TabbedPanelsContentGroup"> | ||
| + | <div class="TabbedPanelsContent"> | ||
| + | |||
| + | |||
| + | <div id="CollapsiblePanelM11" class="CollapsiblePanel"> | ||
| + | <div class="CollapsiblePanelTab" tabindex="0"><h4><i>E. coli</i> breaks down PUR </html><font size="1">▼</font size="1"><html></h4></div> | ||
| + | <div class="CollapsiblePanelContent"> | ||
| + | |||
| + | </html> | ||
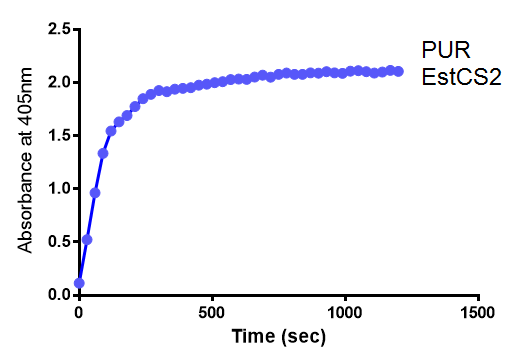
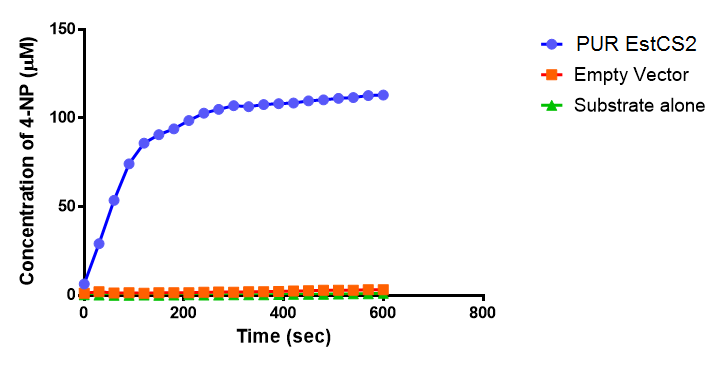
| + | <p>We proved the PUR esterase activity of EstCS2 in colourimetric assay with the substrate analog para-Nitrophenyl butyrate. This result is important because we now have enzyme to break down a common material in mixed plastic waste.</p> | ||
| + | |||
| + | [[File: 4-NP_abs02.png|thumbnail|center|800px|The increase in absorbance that accompanies the cleavage of para-Nitrophenyl butyrate by PUR Esterase EstCS2. Figure by Imperial College London iGEM 2013.]] | ||
| + | [[File: PUREstC2_graph.png|thumbnail|center|700px|The concentration of 4-Nitrophenol released by PUR Esterase EstCS2 activity. Empty Vector and Substrate alone were used as negative controls. Figure by Imperial College London iGEM 2013.]] | ||
| + | |||
| + | |||
| + | <html> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div id="CollapsiblePanelM12" class="CollapsiblePanel"> | ||
| + | <div class="CollapsiblePanelTab" tabindex="0"><h4><i>E. coli</i> grows on ethylene glycol </html><font size="1">▼</font size="1"><html></h4></div> | ||
| + | <div class="CollapsiblePanelContent"> | ||
| + | |||
| + | </html> | ||
| + | <p align="justify>We tested cell growth in ethylene glycol, which is a PUR degradation product. We showed that at 37°C, cell growth is not significantly affected by the concentration of ethylene glycol. Thus our engineered bacteria can grow in the bioreactors!</p> | ||
| + | [[File: EG_growth.png|thumbnail|center|800px|Ethylene glycol in LB with Stress Response cells. <i>E. coli</i>MG1655 were grown in ethylene glycol, a byproduct of polyurethane degradation. Cells were grown in 0mM, 100mM or 200mM Ethylene Glycol. At 37°C, the concentrations of ethylene glycol used do not affect growth, however at 30°C, the increasing concentration results in halved growth. A two-tailed t-test addressing the null hypothesis, temperature does not affect growth with ethylene glycol shows that the null hypothesis must be rejected as p = 0.001. Data points show final time point after 6h growth for each concentration. Growth was at 37°C and 30°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | |||
| + | |||
| + | <html> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div id="CollapsiblePanelM13" class="CollapsiblePanel"> | ||
| + | <div class="CollapsiblePanelTab" tabindex="0"><h4>Modelling </html><font size="1">▼</font size="1"><html></h4></div> | ||
| + | <div class="CollapsiblePanelContent"> | ||
| + | |||
| + | </html> | ||
| + | <p align="justify">Using model simulation results, we predict that the concentration of PhaCAB enzymes </p> | ||
| + | |||
| + | |||
| + | <html> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div id="CollapsiblePanelM14" class="CollapsiblePanel"> | ||
| + | <div class="CollapsiblePanelTab" tabindex="0"><h4>Optimised bioplastic production </html><font size="1">▼</font size="1"><html></h4></div> | ||
| + | <div class="CollapsiblePanelContent"> | ||
| + | |||
| + | </html> | ||
| + | |||
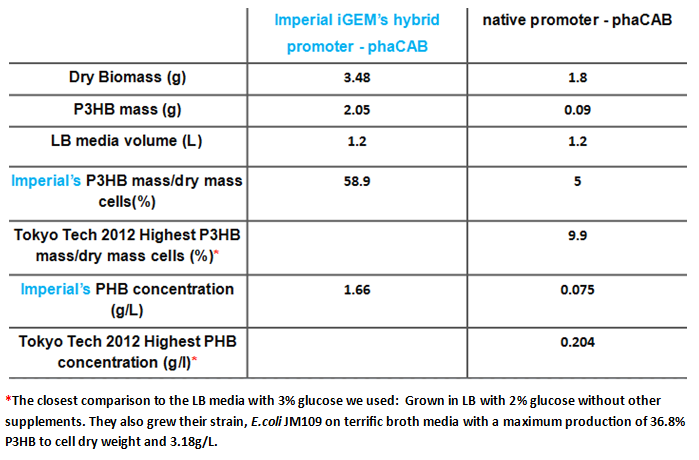
| + | <p align="justify">Since our model predicts that the concentration of PhaB enzymes is the rate limiting step in P3HB production, we designed a [http://parts.igem.org/Part:BBa_K1149051 hybrid promoter] consists of the J23104 constitutive promoter and the native promoter to optimise gene expression. Our results show that we have successfully produced <b>10-fold</b> more P3HB bioplastic compared with the native promoter. </p> | ||
| + | |||
| + | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
| + | |[[File:PHB_production_table.PNG|thumbnail|left|900px| <b>A summary of the improved production of P3HB by our hybrid promoter-phaCAB construct(BBa_K1149051) over the native promoter-phaCAB.</b>]] | ||
| + | |} | ||
| + | |||
| + | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
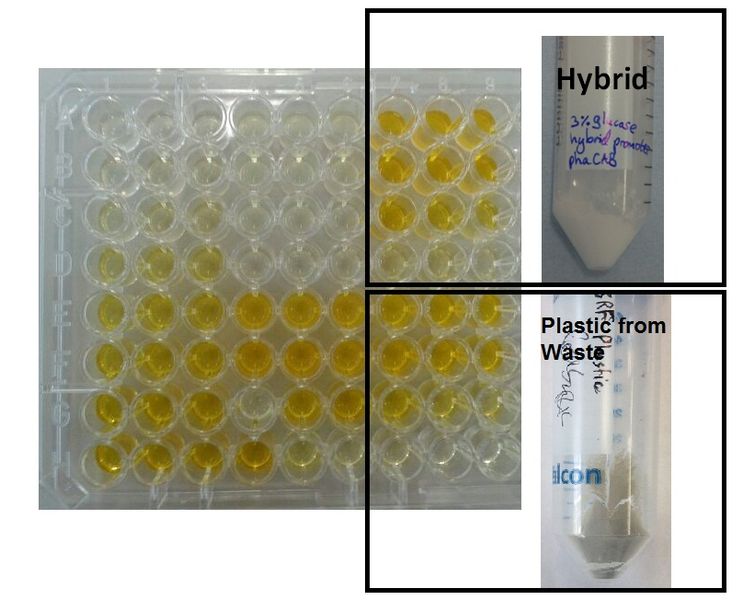
| + | |[[File:Moreplastic.JPG|thumbnail|left|700px|<b>Comparison of P3HB production </b>. <b> (left)</b> 1.5ml tube, natural phaCAB (BBa_K934001) <b>(right)</b> 5ml tube, phaCAB expressed from the hybrid promoter, (BBa_K1149051).]] | ||
| + | |} | ||
| + | |||
| + | <html> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div id="CollapsiblePanelM15" class="CollapsiblePanel"> | ||
| + | <div class="CollapsiblePanelTab" tabindex="0"><h4>Bioplastic from mixed waste </html><font size="1">▼</font size="1"><html></h4></div> | ||
| + | <div class="CollapsiblePanelContent"> | ||
| + | |||
| + | </html> | ||
| + | <p align="justify">One of the objectives of [https://2013.igem.org/Team:Imperial_College/Waste_Degradation:_SRF Module 1] is to produce P3HB bioplastic from waste. By comparing degradation product 3HB of P3HB bought from Sigma, produced from glucose and produced from the waste, we found there is no significant differences in 3HB concentration between these samples.</p> | ||
| + | |||
| + | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
| + | |[[File:743px-3HB_assay_from_PHB_másolata.jpg|thumbnail|left|600px| <b>The chemical analysis of the produced bioplastic. The samples break break down to 3HB monomers after treatment with our PhaZ1 enzyme (BBa_K1149010). We synthesised P(3HB) using our improved Biobrick part (hybrid promoter phaCAB, BBa_K1149051). Our engineered bioplastic producing <i>E.coli</i> synthesised P(3HB) directly from waste. Imperial iGEM data</b>]] | ||
| + | |} | ||
| + | https://static.igem.org/mediawiki/parts/d/d1/3HB_from_PHB_from_waste.jpg | ||
| + | |||
| + | <html> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <h2 id="Plastic Fantastic"></h2> | ||
| + | |||
| + | </div> | ||
| + | <div class="TabbedPanelsContent"> | ||
| + | |||
| + | <div id="CollapsiblePanelM21" class="CollapsiblePanel"> | ||
| + | <div class="CollapsiblePanelTab" tabindex="0"><h4><i>E. coli</i> grows in P3HB and 3HB </html><font size="1">▼</font size="1"><html></h4></div> | ||
| + | <div class="CollapsiblePanelContent">Content | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div id="CollapsiblePanelM22" class="CollapsiblePanel"> | ||
| + | <div class="CollapsiblePanelTab" tabindex="0"><h4><i>E. coli</i> feeds on 3HB </html><font size="1">▼</font size="1"><html></h4></div> | ||
| + | <div class="CollapsiblePanelContent">Content | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div id="CollapsiblePanelM23" class="CollapsiblePanel"> | ||
| + | <div class="CollapsiblePanelTab" tabindex="0"><h4><i>E. coli</i> degrades P3HB </html><font size="1">▼</font size="1"><html></h4></div> | ||
| + | <div class="CollapsiblePanelContent">Content | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div id="CollapsiblePanelM24" class="CollapsiblePanel"> | ||
| + | <div class="CollapsiblePanelTab" tabindex="0"><h4>Modelling P3HB degradation </html><font size="1">▼</font size="1"><html></h4></div> | ||
| + | <div class="CollapsiblePanelContent">Content | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div id="CollapsiblePanelM25" class="CollapsiblePanel"> | ||
| + | <div class="CollapsiblePanelTab" tabindex="0"><h4>PLA degraded by <i>E. coli</i> </html><font size="1">▼</font size="1"><html></h4></div> | ||
| + | <div class="CollapsiblePanelContent">Content | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <script type="text/javascript"> | ||
| + | var TabbedPanels1 = new Spry.Widget.TabbedPanels("TabbedPanels1"); | ||
| + | |||
| + | var CollapsiblePanelM11 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM11", {contentIsOpen:false}); | ||
| + | var CollapsiblePanelM12 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM12", {contentIsOpen:false}); | ||
| + | var CollapsiblePanelM13 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM13", {contentIsOpen:false}); | ||
| + | var CollapsiblePanelM14 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM14", {contentIsOpen:false}); | ||
| + | var CollapsiblePanelM15 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM15", {contentIsOpen:false}); | ||
| + | |||
| + | var CollapsiblePanelM21 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM21", {contentIsOpen:false}); | ||
| + | var CollapsiblePanelM22 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM22", {contentIsOpen:false}); | ||
| + | var CollapsiblePanelM23 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM23", {contentIsOpen:false}); | ||
| + | var CollapsiblePanelM24 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM24", {contentIsOpen:false}); | ||
| + | var CollapsiblePanelM25 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM25", {contentIsOpen:false}); | ||
| + | |||
| + | </script> | ||
| + | <html> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | </html> | ||
{{:Team:Imperial_College/Templates:footer}} | {{:Team:Imperial_College/Templates:footer}} | ||
Revision as of 16:33, 4 October 2013
Main Results
Resource-full Waste
Plastic Fantastic
E. coli breaks down PUR ▼
We proved the PUR esterase activity of EstCS2 in colourimetric assay with the substrate analog para-Nitrophenyl butyrate. This result is important because we now have enzyme to break down a common material in mixed plastic waste.
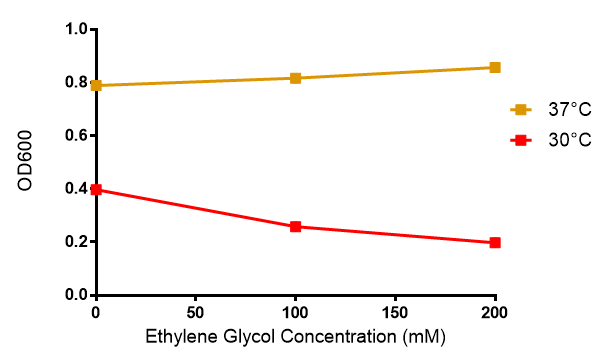
E. coli grows on ethylene glycol ▼
We tested cell growth in ethylene glycol, which is a PUR degradation product. We showed that at 37°C, cell growth is not significantly affected by the concentration of ethylene glycol. Thus our engineered bacteria can grow in the bioreactors!
Modelling ▼
Using model simulation results, we predict that the concentration of PhaCAB enzymes
Optimised bioplastic production ▼
Since our model predicts that the concentration of PhaB enzymes is the rate limiting step in P3HB production, we designed a [http://parts.igem.org/Part:BBa_K1149051 hybrid promoter] consists of the J23104 constitutive promoter and the native promoter to optimise gene expression. Our results show that we have successfully produced 10-fold more P3HB bioplastic compared with the native promoter.
Bioplastic from mixed waste ▼
One of the objectives of Module 1 is to produce P3HB bioplastic from waste. By comparing degradation product 3HB of P3HB bought from Sigma, produced from glucose and produced from the waste, we found there is no significant differences in 3HB concentration between these samples.

E. coli grows in P3HB and 3HB ▼
E. coli feeds on 3HB ▼
E. coli degrades P3HB ▼
Modelling P3HB degradation ▼
PLA degraded by E. coli ▼
 "
"