Team:Heidelberg/Tyrocidine week17 interms
From 2013.igem.org
(Created page with " ==Amplifications == In the course of this project, we'll have to amplify the following fragments: [[Media:Fragments Tyrocidine-Indigoidine-Fusion.pdf|Fragments for Tyrocidine-...") |
|||
| Line 1: | Line 1: | ||
| - | + | ||
==Amplifications == | ==Amplifications == | ||
| Line 7: | Line 7: | ||
For reasons of clarity, the gel-pictures obtained by PCR are shown here in this gallery: | For reasons of clarity, the gel-pictures obtained by PCR are shown here in this gallery: | ||
| - | [[File: | + | [[File:Heidelberg_A-I 1st PCR.png|300px]] |
| - | [[File: | + | [[File:Heidelberg_AEG 2nd PCR.png|150px]] |
| - | [[File: | + | [[File:Heidelberg_A-I 3rd PCR.png|300px]] |
| - | [[File: | + | [[File:Heidelberg_D 4th PCR.png|300px]] |
| - | Comment on the second | + | Comment on the second File:Heidelberg_ |
The normal gel-image before the cut out was not properly saved on the PC that is connected to the camera. The bands that were visible (one can estimate them by the cut-out) were at about 2.4 to 2.5 kbp, which is the expected size of the backbone | The normal gel-image before the cut out was not properly saved on the PC that is connected to the camera. The bands that were visible (one can estimate them by the cut-out) were at about 2.4 to 2.5 kbp, which is the expected size of the backbone | ||
| Line 18: | Line 18: | ||
=== Fragment A === | === Fragment A === | ||
==== A ==== | ==== A ==== | ||
| - | [[File:...]] | + | [[File:Heidelberg_...]] |
{| class="wikitable" | {| class="wikitable" | ||
| Line 136: | Line 136: | ||
=== Fragment B === | === Fragment B === | ||
==== A ==== | ==== A ==== | ||
| - | [[File:...]] | + | [[File:Heidelberg_...]] |
{| class="wikitable" | {| class="wikitable" | ||
| Line 215: | Line 215: | ||
=== Fragment C === | === Fragment C === | ||
==== A ==== | ==== A ==== | ||
| - | [[File:...]] | + | [[File:Heidelberg_...]] |
{| class="wikitable" | {| class="wikitable" | ||
| Line 340: | Line 340: | ||
=== Fragment E === | === Fragment E === | ||
==== A ==== | ==== A ==== | ||
| - | [[File:...]] | + | [[File:Heidelberg_...]] |
{| class="wikitable" | {| class="wikitable" | ||
| Line 458: | Line 458: | ||
=== Fragment F === | === Fragment F === | ||
==== A ==== | ==== A ==== | ||
| - | [[File:...]] | + | [[File:Heidelberg_...]] |
{| class="wikitable" | {| class="wikitable" | ||
| Line 537: | Line 537: | ||
=== Fragment G === | === Fragment G === | ||
==== A ==== | ==== A ==== | ||
| - | [[File:...]] | + | [[File:Heidelberg_...]] |
{| class="wikitable" | {| class="wikitable" | ||
| Line 655: | Line 655: | ||
=== Fragment H === | === Fragment H === | ||
==== A ==== | ==== A ==== | ||
| - | [[File:...]] | + | [[File:Heidelberg_...]] |
{| class="wikitable" | {| class="wikitable" | ||
| Line 734: | Line 734: | ||
=== Fragment I === | === Fragment I === | ||
==== A ==== | ==== A ==== | ||
| - | [[File:...]] | + | [[File:Heidelberg_...]] |
{| class="wikitable" | {| class="wikitable" | ||
| Line 816: | Line 816: | ||
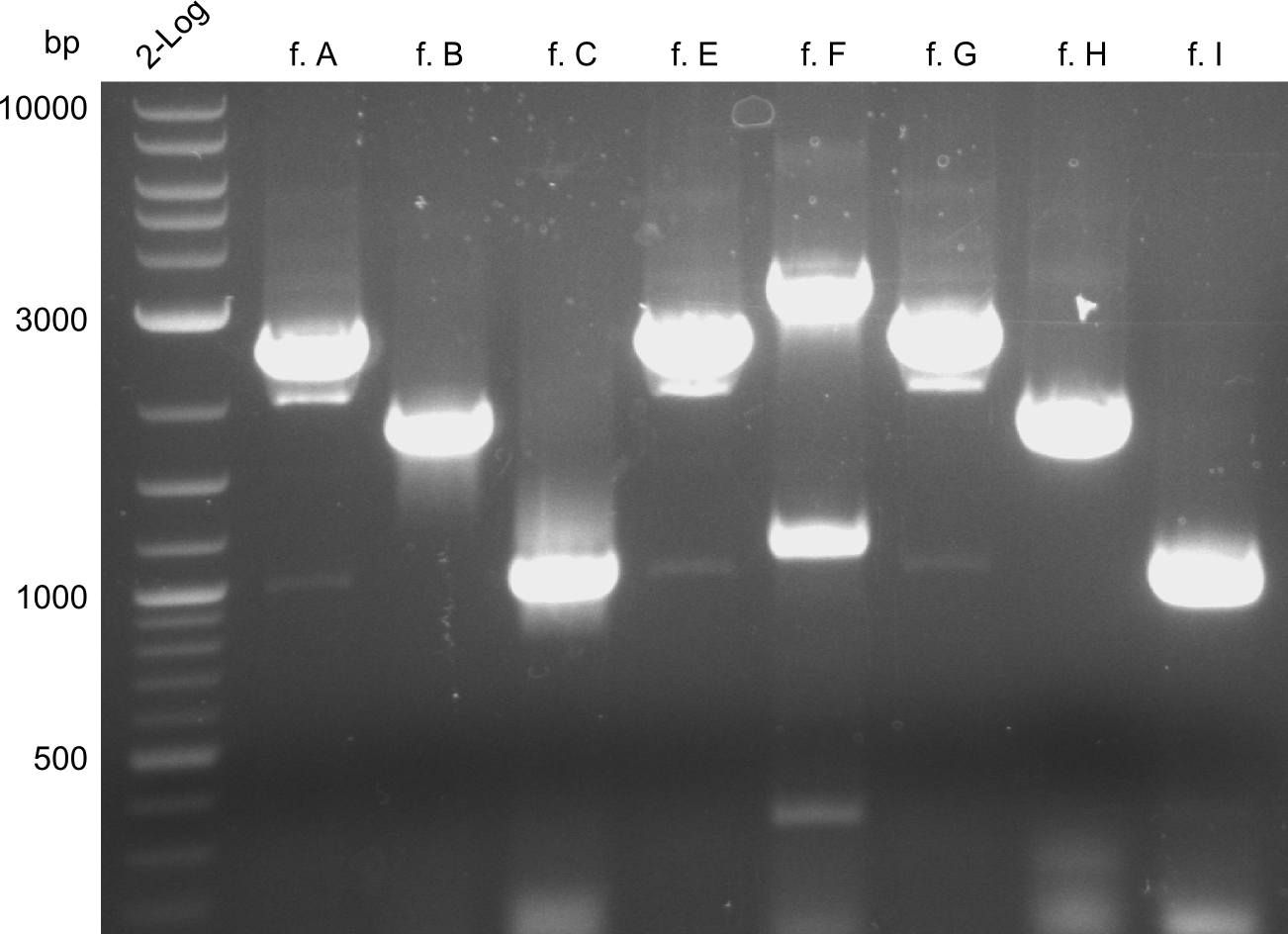
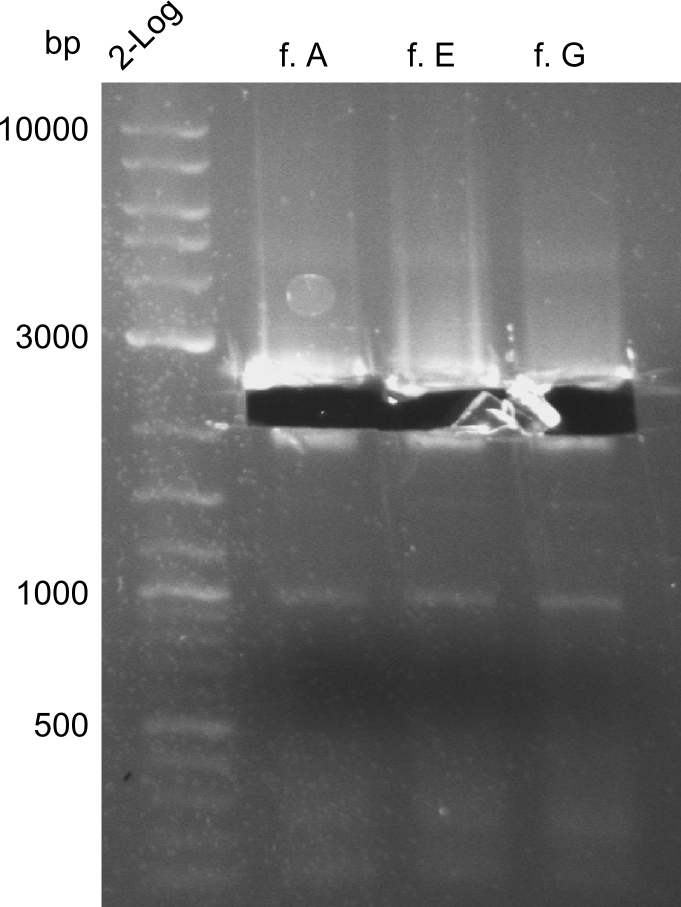
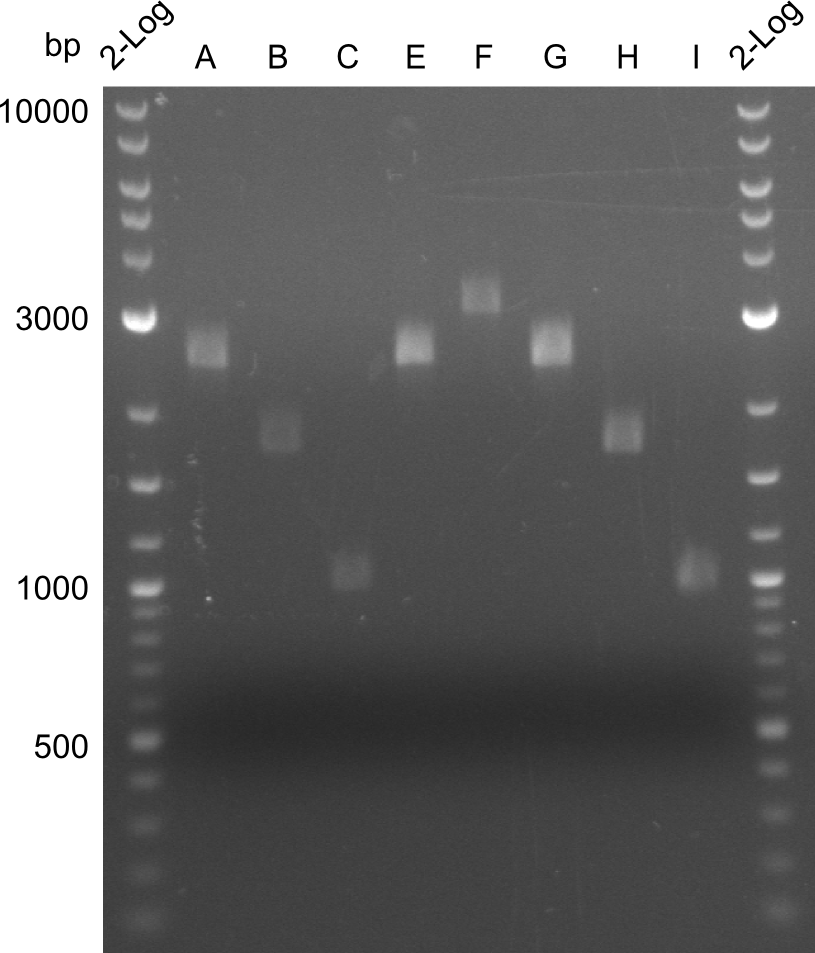
The fragments A, E and G from the first PCR were not used due to the big smear and the double-bands, instead the bands obtained in the second PCR were taken for the quantification. | The fragments A, E and G from the first PCR were not used due to the big smear and the double-bands, instead the bands obtained in the second PCR were taken for the quantification. | ||
| - | [[File: | + | [[File:Heidelberg_A-I 1st quant.png|300px]] |
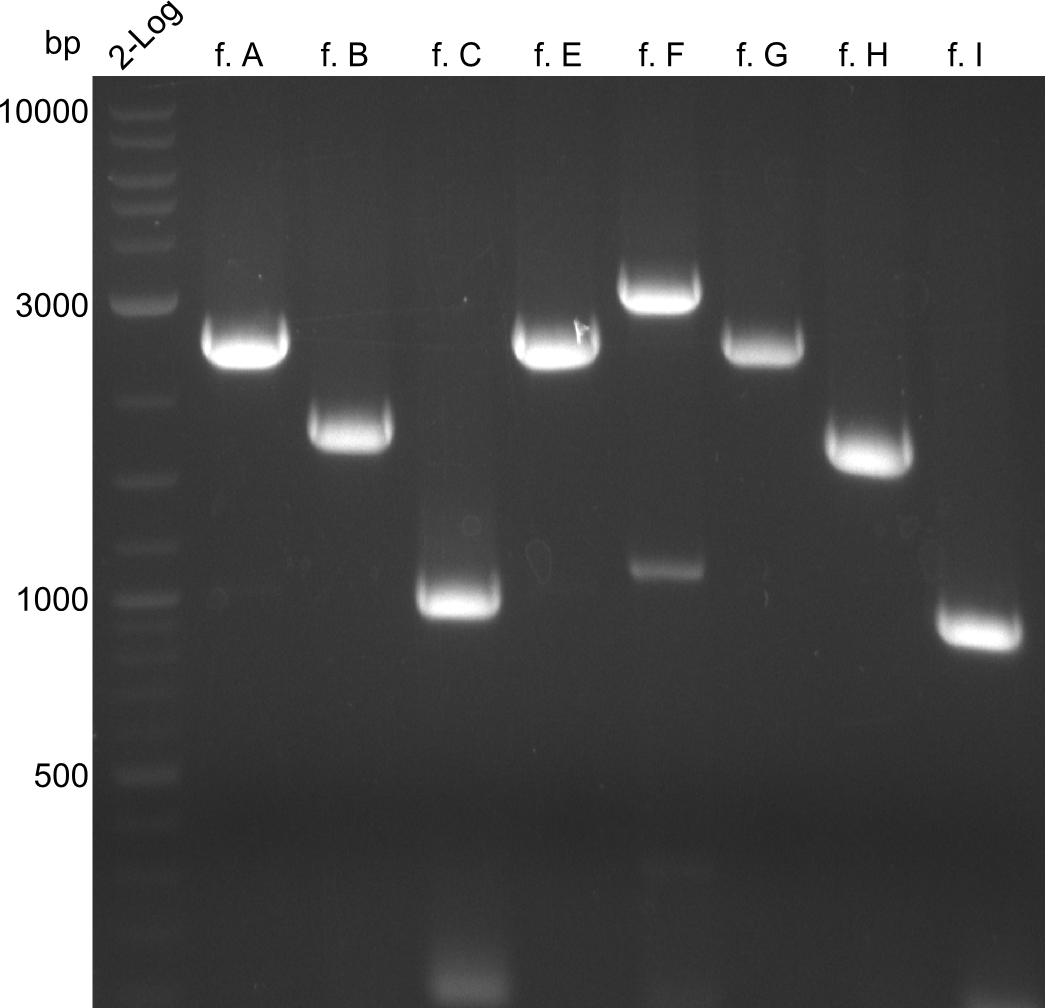
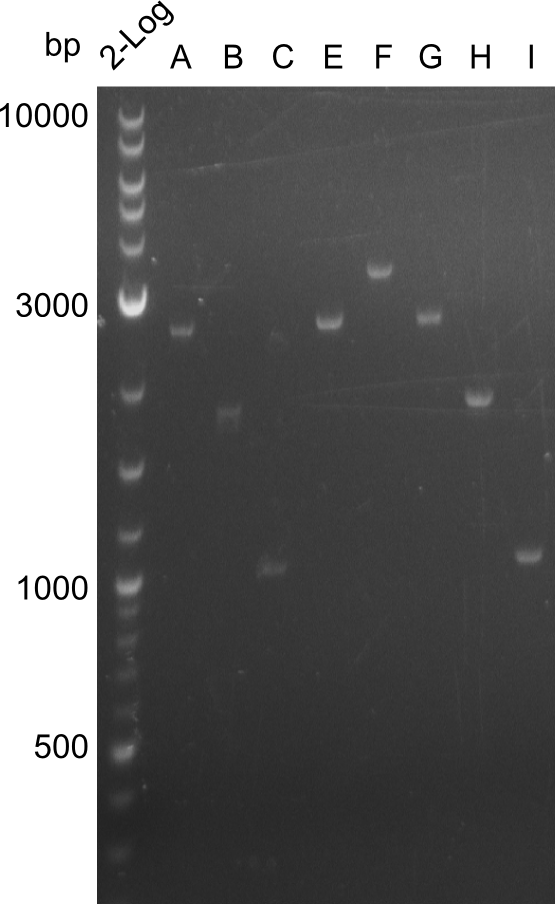
The bands did not have a definite size, hence they are not useful to work with due to unwanted byproducts that are obviously present. After the optimizations regarding annealing-temperature and cycle-repetition, the following bands were obtained during quantification | The bands did not have a definite size, hence they are not useful to work with due to unwanted byproducts that are obviously present. After the optimizations regarding annealing-temperature and cycle-repetition, the following bands were obtained during quantification | ||
| - | [[File: | + | [[File:Heidelberg_A-I 2nd quant.png|300px]] |
| - | [[File: | + | [[File:Heidelberg_D 3rd quant.png|122px]] |
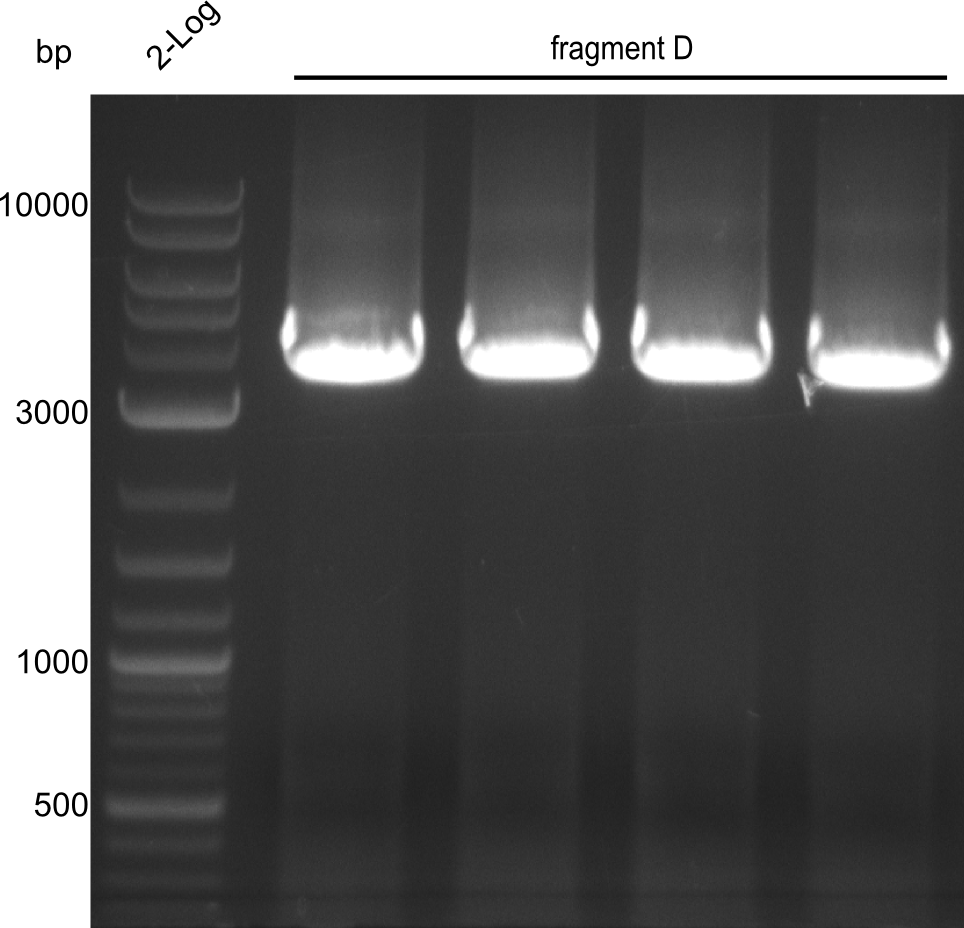
Here, the bands have a definded size and a sufficient quantity to work with. The quantification for fragment D was carried out later, as the amplification of those fragments happened later. | Here, the bands have a definded size and a sufficient quantity to work with. The quantification for fragment D was carried out later, as the amplification of those fragments happened later. | ||
Latest revision as of 16:40, 4 October 2013
Contents |
Amplifications
In the course of this project, we'll have to amplify the following fragments: Fragments for Tyrocidine-Indigoidine-Fusion
For reasons of clarity, the gel-pictures obtained by PCR are shown here in this gallery:
Comment on the second File:Heidelberg_
The normal gel-image before the cut out was not properly saved on the PC that is connected to the camera. The bands that were visible (one can estimate them by the cut-out) were at about 2.4 to 2.5 kbp, which is the expected size of the backbone
Fragment A
A
| what | µl |
|---|---|
| pSB1C3 | 1 |
| IK21 | 2 |
| PW23 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 35 | 98 | 1 |
| 66 | 5 | |
| 72 | 40 | |
| 1 | 72 | 300 |
| 1 | 10 | inf |
Results
The gel was overloaded, which might be due to a too low dilution of Medi-Prep and hence too much template. For optimization use less template and take it from a more diluted sample.
B
| what | µl |
|---|---|
| pSB1C3 | 0.5 |
| IK21 | 2 |
| PW23 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5.5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 35 | 98 | 1 |
| 66 | 5 | |
| 72 | 40 | |
| 1 | 72 | 300 |
| 1 | 10 | inf |
Results
Still the gel is overloaded, though less template was used. For the next time, reduce cycle repetitin from 35 to 30 and increase annealing temperature by 1°C
C
| what | µl |
|---|---|
| pSB1C3 | 0.5 |
| IK21 | 2 |
| PW23 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5.5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 67 | 5 | |
| 72 | 40 | |
| 1 | 72 | 300 |
| 1 | 10 | inf |
Results
Fragment B
A
| what | µl |
|---|---|
| B. parabrevis | 1 |
| IK16 | 2 |
| PW17 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 120 |
| 35 | 98 | 1 |
| 65 | 5 | |
| 72 | 40 | |
| 1 | 72 | 300 |
| 1 | 10 | inf |
Results
The gel was overloaded, which might be due to the polymerase, as Q5 was usually used in previous samples and bright bands are visible for all fragments. For optimization, the annealing-temperature could be increased by 1 - 2°C and cycle repetition may be reduced to 30 cycles.
B
| what | µl |
|---|---|
| B. parabrevis | 1 |
| IK16 | 2 |
| PW17 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 120 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 40 | |
| 1 | 72 | 300 |
| 1 | 10 | inf |
Results
Fragment C
A
| what | µl |
|---|---|
| B. parabrevis | 1 |
| PW18 | 2 |
| PW14 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 120 |
| 35 | 98 | 1 |
| 65 | 5 | |
| 72 | 40 | |
| 1 | 72 | 300 |
| 1 | 10 | inf |
Results
The gel was overloaded, which might be due to the polymerase, as Q5 was usually used in previous samples and bright bands are visible for all fragments. For optimization, the annealing-temperature could be increased by 1 - 2°C and cycle repetition may be reduced to 30 cycles.
B
| what | µl |
|---|---|
| B. parabrevis | 1 |
| PW18 | 2 |
| PW14 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 120 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 40 | |
| 1 | 72 | 300 |
| 1 | 10 | inf |
Results
Fragment D
A
For obtaining more product and optimizing the conditions simultaneously, a gradient PCR with touch-down was carried out. 4 wells were used at 62°C, 61.4°C, 60.6°C and 60°C for touchdown, with steady temperatures at 59°C, 58.4°C, 57.6°C and 57°C.
| what | µl |
|---|---|
| indC (RB37, RB38) | 0.5 |
| PW15 | 2 |
| PW16 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5.5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 5 |
| 62 ↓ 0.5 to 60 ↓ 0.5 | 5 | |
| 72 | 100 | |
| 23 | 98 | 5 |
| 59 to 57 | 10 | |
| 72 | 100 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
Results
Fragment E
A
| what | µl |
|---|---|
| pSB1C3 | 1 |
| PW25 | 2 |
| PW23 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 35 | 98 | 1 |
| 66 | 5 | |
| 72 | 40 | |
| 1 | 72 | 300 |
| 1 | 10 | inf |
Results
The gel was overloaded, which might be due to a too low dilution of Medi-Prep and hence too much template. For optimization use less template and take it from a more diluted sample.
B
| what | µl |
|---|---|
| pSB1C3 | 0.5 |
| PW25 | 2 |
| PW23 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5.5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 35 | 98 | 1 |
| 66 | 5 | |
| 72 | 40 | |
| 1 | 72 | 300 |
| 1 | 10 | inf |
Result
Still the gel is overloaded, though less template was used. For the next time, reduce cycle repetitin from 35 to 30 and increase annealing temperature by 1°C
C
| what | µl |
|---|---|
| pSB1C3 | 0.5 |
| PW25 | 2 |
| PW23 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5.5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 67 | 5 | |
| 72 | 40 | |
| 1 | 72 | 300 |
| 1 | 10 | inf |
Results
Fragment F
A
| what | µl |
|---|---|
| B. parabrevis | 1 |
| PW24 | 2 |
| PW14 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 120 |
| 35 | 98 | 1 |
| 59 | 5 | |
| 72 | 60 | |
| 1 | 72 | 300 |
| 1 | 10 | inf |
Results
The gel was overloaded, which might be due to the polymerase, as Q5 was usually used in previous samples and bright bands are visible for all fragments. For optimization, the annealing-temperature could be increased by 1 - 2°C and cycle repetition may be reduced to 30 cycles.
B
| what | µl |
|---|---|
| B. parabrevis | 1 |
| PW24 | 2 |
| PW14 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 120 |
| 30 | 98 | 1 |
| 61 | 5 | |
| 72 | 60 | |
| 1 | 72 | 300 |
| 1 | 10 | inf |
Results
Fragment G
A
| what | µl |
|---|---|
| pSB1C3 | 1 |
| PW22 | 2 |
| PW23 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 35 | 98 | 1 |
| 66 | 5 | |
| 72 | 40 | |
| 1 | 72 | 300 |
| 1 | 10 | inf |
Results
The gel was overloaded, which might be due to a too low dilution of Medi-Prep and hence too much template. For optimization use less template and take it from a more diluted sample.
B
| what | µl |
|---|---|
| pSB1C3 | 0.5 |
| PW22 | 2 |
| PW23 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5.5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 35 | 98 | 1 |
| 66 | 5 | |
| 72 | 40 | |
| 1 | 72 | 300 |
| 1 | 10 | inf |
Results
Still the gel is overloaded, though less template was used. For the next time, reduce cycle repetitin from 35 to 30 and increase annealing temperature by 1°C
C
| what | µl |
|---|---|
| pSB1C3 | 0.5 |
| PW22 | 2 |
| PW23 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5.5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 67 | 5 | |
| 72 | 40 | |
| 1 | 72 | 300 |
| 1 | 10 | inf |
Results
Fragment H
A
| what | µl |
|---|---|
| B. parabrevis | 1 |
| PW19 | 2 |
| PW20 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 120 |
| 35 | 98 | 1 |
| 59 | 5 | |
| 72 | 60 | |
| 1 | 72 | 300 |
| 1 | 10 | inf |
Results
The gel was overloaded, which might be due to the polymerase, as Q5 was usually used in previous samples and bright bands are visible for all fragments. For optimization, the annealing-temperature could be increased by 1 - 2°C and cycle repetition may be reduced to 30 cycles.
B
| what | µl |
|---|---|
| B. parabrevis | 1 |
| PW19 | 2 |
| PW20 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 120 |
| 30 | 98 | 1 |
| 61 | 5 | |
| 72 | 60 | |
| 1 | 72 | 300 |
| 1 | 10 | inf |
Results
Fragment I
A
| what | µl |
|---|---|
| B. parabrevis | 1 |
| PW21 | 2 |
| PW14 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 120 |
| 35 | 98 | 1 |
| 59 | 5 | |
| 72 | 60 | |
| 1 | 72 | 300 |
| 1 | 10 | inf |
Results
The gel was overloaded, which might be due to the polymerase, as Q5 was usually used in previous samples and bright bands are visible for all fragments. For optimization, the annealing-temperature could be increased by 1 - 2°C and cycle repetition may be reduced to 30 cycles.
B
| what | µl |
|---|---|
| B. parabrevis | 1 |
| PW21 | 2 |
| PW14 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 120 |
| 30 | 98 | 1 |
| 61 | 5 | |
| 72 | 60 | |
| 1 | 72 | 300 |
| 1 | 10 | inf |
Results
Quantification-gels
For the further work, the amount of DNA available has to be quantified. We decided to do this by a quantification gel in order to check the right sizes of the constructs. The fragments A, E and G from the first PCR were not used due to the big smear and the double-bands, instead the bands obtained in the second PCR were taken for the quantification.
The bands did not have a definite size, hence they are not useful to work with due to unwanted byproducts that are obviously present. After the optimizations regarding annealing-temperature and cycle-repetition, the following bands were obtained during quantification
Here, the bands have a definded size and a sufficient quantity to work with. The quantification for fragment D was carried out later, as the amplification of those fragments happened later.
 "
"