Team:Heidelberg/Tyrocidine week21 variation
From 2013.igem.org
(Created page with " To design the Linker Variation Constructs, we amplified the DNA fragments LV1a, LV1b, LV2b, LV3a, LV5b and LV6 by PCR with the following protocol. ...") |
|||
| Line 1: | Line 1: | ||
| - | |||
To design the [[Media:Construct_Table.pdf|Linker Variation Constructs]], we amplified the DNA fragments LV1a, LV1b, LV2b, LV3a, LV5b and LV6 by PCR with the following protocol. | To design the [[Media:Construct_Table.pdf|Linker Variation Constructs]], we amplified the DNA fragments LV1a, LV1b, LV2b, LV3a, LV5b and LV6 by PCR with the following protocol. | ||
(Primer are listed on our [[Primer#Linker_Variation|primer-page]]) | (Primer are listed on our [[Primer#Linker_Variation|primer-page]]) | ||
| Line 54: | Line 53: | ||
| 1|| 10 || inf | | 1|| 10 || inf | ||
|} | |} | ||
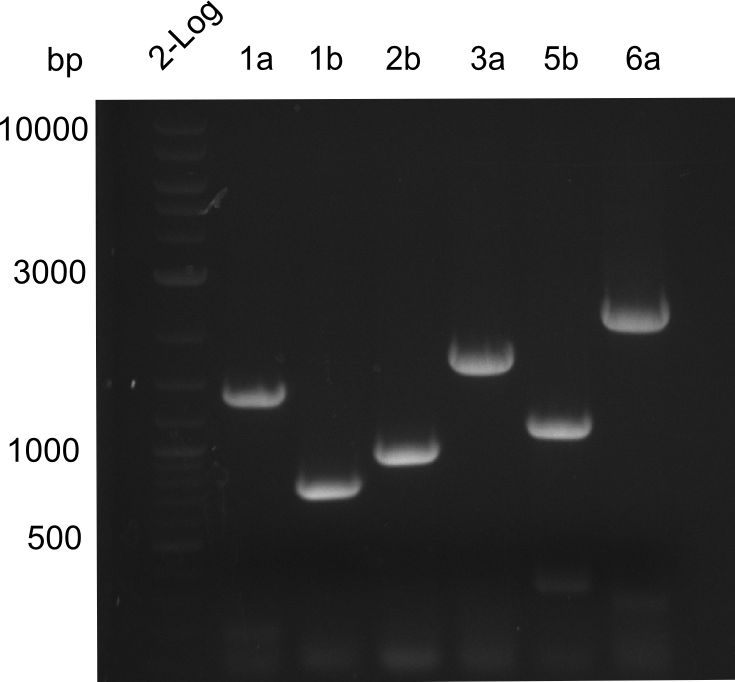
| - | [[File: | + | [[File:Heidelberg_20130919_amplification1-6.png|right|100px|thumb|Linker variation constructs 1-6 after PCR]] |
===Results=== | ===Results=== | ||
We were able to amplify all of our constructs, each of them run on the expected size. | We were able to amplify all of our constructs, each of them run on the expected size. | ||
| Line 120: | Line 119: | ||
As we were able to amplify all af the constructs, we quantified the gained concentrations for gibson assembly. | As we were able to amplify all af the constructs, we quantified the gained concentrations for gibson assembly. | ||
<gallery> | <gallery> | ||
| - | File: | + | File:Heidelberg_20130920_amplificationbb+ind.png|Constructs of bb and indC after PCR. |
| - | File: | + | File:Heidelberg_20130919_20130922_quantificationbb+ind.png|Concentrations of bb- and indC-constructs. |
</gallery> | </gallery> | ||
{| class="wikitable" | {| class="wikitable" | ||
Latest revision as of 17:15, 4 October 2013
To design the Linker Variation Constructs, we amplified the DNA fragments LV1a, LV1b, LV2b, LV3a, LV5b and LV6 by PCR with the following protocol. (Primer are listed on our primer-page)
| What | µl |
|---|---|
| primer_fw (see link above) | 2 |
| primer_rv (see link above) | 2 |
| Brevibacillus parabrevis | 1 |
| Phusion Flash 2x Master Mix | 10 |
| ddH20 | 5 |
- PCR-Program LV1a, LV1b, LV2b, LV5b amplification
| Cycles | Temperature [°C] | Time [min:s] |
|---|---|---|
| 1 | 98 | 2:00 |
| 35 | 98 | 0:02 |
| 59,4 | 0:05 | |
| 72 | 0:28 | |
| 1 | 72 | 10:00 |
| 1 | 10 | inf |
- PCR-Program LV3a, LV6a amplification
| Cycles | Temperature [°C] | Time [min:s] |
|---|---|---|
| 1 | 98 | 2:00 |
| 35 | 98 | 0:02 |
| 59,4 | 0:05 | |
| 72 | 0:44 | |
| 1 | 72 | 10:00 |
| 1 | 10 | inf |
Contents |
Results
We were able to amplify all of our constructs, each of them run on the expected size.
Gel extraction
The amplified constructs LV14,LV1b,LV2b,LV3a,LV5b,LV6a were sliced out of the gel and a gel extraction performed. Afterwards, the concentration was measured with a nanodrop.
Results
| Construct | Concentration in ng/µl | A280/A260 |
|---|---|---|
| LV1a | 41,6 | 1,89 |
| LV1b | 69,4 | 1,86 |
| LV2b | 74,4 | 1,97 |
| LV3a | 72,9 | 1,84 |
| LV5b | 56,7 | 1,94 |
| LV6a | 50,7 | 2,05 |
Amplification of Backbone and indC
As we want to usethe strategy we established to detect a successful transformation by the expression of indigoidine, we amplified not only the backbone, but also the indigoidine constructs. They were amplified with both, a gibson-overhang for the short-linker-constructs and the long-linker-constructs.
- PCR-Program backbone short- and long-linker amplification
| Cycles | Temperature [°C] | Time [min:s] |
|---|---|---|
| 1 | 98 | 0:05 |
| 35 | 98 | 0:02 |
| 66,2 | 0:05 | |
| 72 | 0:50 | |
| 1 | 72 | 10:00 |
| 1 | 10 | inf |
- PCR-Program indigoidine short- and long-linker amplification
| Cycles | Temperature [°C] | Time [min:s] |
|---|---|---|
| 1 | 98 | 0:05 |
| 35 | 98 | 0:02 |
| 56,1 | 0:05 | |
| 72 | 1:20 | |
| 1 | 72 | 10:00 |
| 1 | 10 | inf |
Results
As we were able to amplify all af the constructs, we quantified the gained concentrations for gibson assembly.
| Construct | Concentration ind ng/µl |
|---|---|
| BBs | 10 |
| BBl | 40 |
| Inds | 45 |
| Indl | 45 |
 "
"