Team:Paris Saclay/Notebook/August/30
From 2013.igem.org
(→Notebook : August 30) |
(→6 - Transformation of ligation of Pfnr with RBS-LacZ-Term in pSB3K3 and PnarK with RBS-LacZ-Term in pSB3K3 in DH5α strain) |
||
| Line 73: | Line 73: | ||
Protocol : [[Team:Paris_Saclay/Protocols/Transformation|Bacterial transformation]] | Protocol : [[Team:Paris_Saclay/Protocols/Transformation|Bacterial transformation]] | ||
| - | We incubate the bacteria at 37°C with LB-Kanamycine-Xgal in anaerobic conditions for PnarK with RBS-LacZ-Term in | + | We incubate the bacteria at 37°C with LB-Kanamycine-Xgal in anaerobic conditions for PnarK with RBS-LacZ-Term in pSB3K3. |
We incubate the bacteria at 37°C with LB-Kanamycine-Xgal in aerobic conditions for Pndh* with RBS-LacZ-Term in pSB3K3. | We incubate the bacteria at 37°C with LB-Kanamycine-Xgal in aerobic conditions for Pndh* with RBS-LacZ-Term in pSB3K3. | ||
Revision as of 17:39, 4 October 2013
Notebook : August 30
Lab work
A - Aerobic/Anaerobic regulation system
Objective : characterize BBa_K1155000, BBa_K1155004, BBa_K1155005, BBa_K1155006
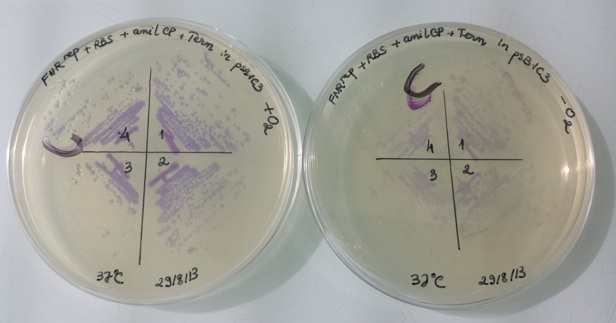
1 - Results of liquid culture of Pfnr-RBS-Amil CP-Term in pSB1C3 in aerobic and anaerobic conditions
XiaoJing
|
IT WORKS !!! |
In anaerobic condition, FNR protein (active form) binds to the constitutive promoter Pndh* and repressed amilCP expression. Therefore, the bacteria have no colour.
In aerobic condition, FNR protein is inactive and can not bind to constitutive promoter Pndh*, amilCP is expressed and we can see the bacteria have violet colour.
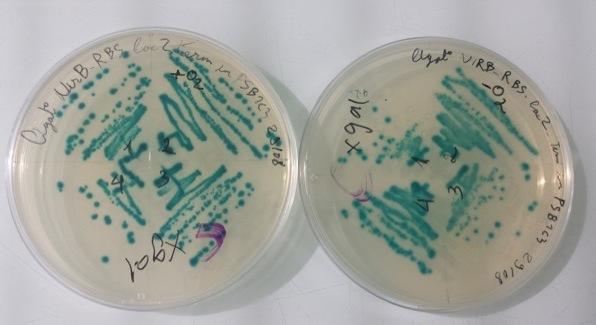
2 - Results of culture of Pfnr with RBS-AmilCP-Term in pSB1C3, PnirB with RBS-LacZ-Term in pSB1C3 in aerobic and anaerobic conditions
XiaoJing
|
Purification of 08/29/13 didn't work. We have blue colonies for Pndh* with RBS-AmilCP-Term in pSB1C3 in aerobic and anaerobic conditions. We also have blue colonies for PnirB with RBS-LacZ-Term in pSB1C3 in anaerobic conditions. |
3 - Ligation of Pndh* with RBS-LacZ-Term in pSB1C3
XiaoJing
Used quantities :
- Pndh* : 8µL
- RBS-LacZ-Term in pSB1C3 : 3µL
- Buffer ligation : 2µL
- Ligase : 1µL
- H2O : 6µL
4 - Transformation of ligation of Pndh* with RBS-LacZ-Term in pSB1C3 in DH5α strain
XiaoJing
Protocol : Bacterial transformation
We incubate the bacteria in LB-chloramphenicol-Xgal plate at 37°C in aerobic conditions.
5 - Ligation of Pfnr with RBS-LacZ-Term in pSB3K3 and PnarK with RBS-LacZ-Term in pSB3K3
XiaoJing
Used quantities :
- Pndh*, PnarK : 3µL
- RBS-LacZ-Term : 3µL
- pSB3K3 : 5µL
- Buffer ligase : 2µL
- Ligase : 1µL
- H2O : 6µL
6 - Transformation of ligation of Pfnr with RBS-LacZ-Term in pSB3K3 and PnarK with RBS-LacZ-Term in pSB3K3 in DH5α strain
XiaoJing
Protocol : Bacterial transformation
We incubate the bacteria at 37°C with LB-Kanamycine-Xgal in anaerobic conditions for PnarK with RBS-LacZ-Term in pSB3K3.
We incubate the bacteria at 37°C with LB-Kanamycine-Xgal in aerobic conditions for Pndh* with RBS-LacZ-Term in pSB3K3.
7 - Electrophoresis of PCR Colony of Pfnr with RBS_AmilCP-Term in DH5α strain
XiaoJing
| [[]] |
|
Expected sizes :
- Pfnr with RBS_AmilCP-Term : 1000bp
|
We obtain fragments at the right size. |
A - Aerobic/Anaerobic regulation system / B - PCB sensor system
Objective : obtaining FNR and BphR2 proteins
1 - Electrophoresis of PCR Colony of FNR, RBS-FNR and RBS-BphR2
| ]] |
|
Expect sizes :
- FNR : 1096 bp
- RBS-FNR : 1014 bp
- RBS-BphR2 : 1469 bp
|
We obtain fragment at right size for FNR and RBS-FNR. The Gisbon assembly of 08/26/13 was good. |
| Previous day | Back to calendar | Next day |
 "
"