Team:Heidelberg/Templates/Del week11 overview
From 2013.igem.org
(Created page with " ==Strategy== pFSN plasmid including Primers FS_01 to FS_16 us...") |
|||
| (4 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
==Strategy== | ==Strategy== | ||
| - | [[File:Heidelberg_Strategie2.png| | + | [[File:Heidelberg_Strategie2.png|300px|right|thumb|Vector map of [[:File:Heidelberg_Psb4k5+BBa J04450 DelRest.gb| pFSN plasmid]] including Primers FS_01 to FS_16 used for assembly of the desired genes from the ''D. acidovorans'' genome as well as the partsregistry backbone pSB4K5 .]] |
We had various problems with the amplification of the fragments located in theregion DelF-G and DelO-P. For these particular PCRs long primers containing the Gibson overlaps had to be used. As these long primers often form complex secondary structures decreasing annealing efficiency we designed short version of these primers, having the same sequence region complementary to the genomic target, but lacking any longer overhangs. Consequently these primesrs will serve only for amplification of the desired genes from the genome of ''D. Acidovorans'' to obtain specific templates. This specific template afterwards will be used to introduce the designated overlap using the long primers again. In total 3 short primer versions FS_11s, FS_13s and FS_15s were ordered. | We had various problems with the amplification of the fragments located in theregion DelF-G and DelO-P. For these particular PCRs long primers containing the Gibson overlaps had to be used. As these long primers often form complex secondary structures decreasing annealing efficiency we designed short version of these primers, having the same sequence region complementary to the genomic target, but lacking any longer overhangs. Consequently these primesrs will serve only for amplification of the desired genes from the genome of ''D. Acidovorans'' to obtain specific templates. This specific template afterwards will be used to introduce the designated overlap using the long primers again. In total 3 short primer versions FS_11s, FS_13s and FS_15s were ordered. | ||
| Line 9: | Line 9: | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| - | + | ====Primer pairs, corresponding sequences and usage==== | |
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
| Line 22: | Line 22: | ||
==Amplification of Del Genes from DSM-39 genome== | ==Amplification of Del Genes from DSM-39 genome== | ||
| - | ===Goals=== | + | ====Goals==== |
The PCRs which worked in the previous week will be repeated to obtain higher product yields, as high concentrations of the long amplicons are needed for a successfull Gibson Assembly. | The PCRs which worked in the previous week will be repeated to obtain higher product yields, as high concentrations of the long amplicons are needed for a successfull Gibson Assembly. | ||
Furthermore we will try to establish PCRs for the obtained short Primers and thereby provide specific templates for the second amplification step using the corresponding primers introducing the required Gibson overlaps. | Furthermore we will try to establish PCRs for the obtained short Primers and thereby provide specific templates for the second amplification step using the corresponding primers introducing the required Gibson overlaps. | ||
| Line 29: | Line 29: | ||
As the amplification of DelO-P also turned out to be difficult, we decided to try a backup strategy which not only includes the desired genes DelO and DelP but also DelL and, since they are located between DelP and DelL, DelM and DelN. These two genes would increase our amplicon size by only 2.2 kb, but we hoped that on the other hand the corresponding primers would work better.<br/> | As the amplification of DelO-P also turned out to be difficult, we decided to try a backup strategy which not only includes the desired genes DelO and DelP but also DelL and, since they are located between DelP and DelL, DelM and DelN. These two genes would increase our amplicon size by only 2.2 kb, but we hoped that on the other hand the corresponding primers would work better.<br/> | ||
| - | ===Results=== | + | ====Results==== |
| - | + | ||
{| class="wikitable" | {| class="wikitable" | ||
Latest revision as of 18:21, 4 October 2013
Contents |
Strategy
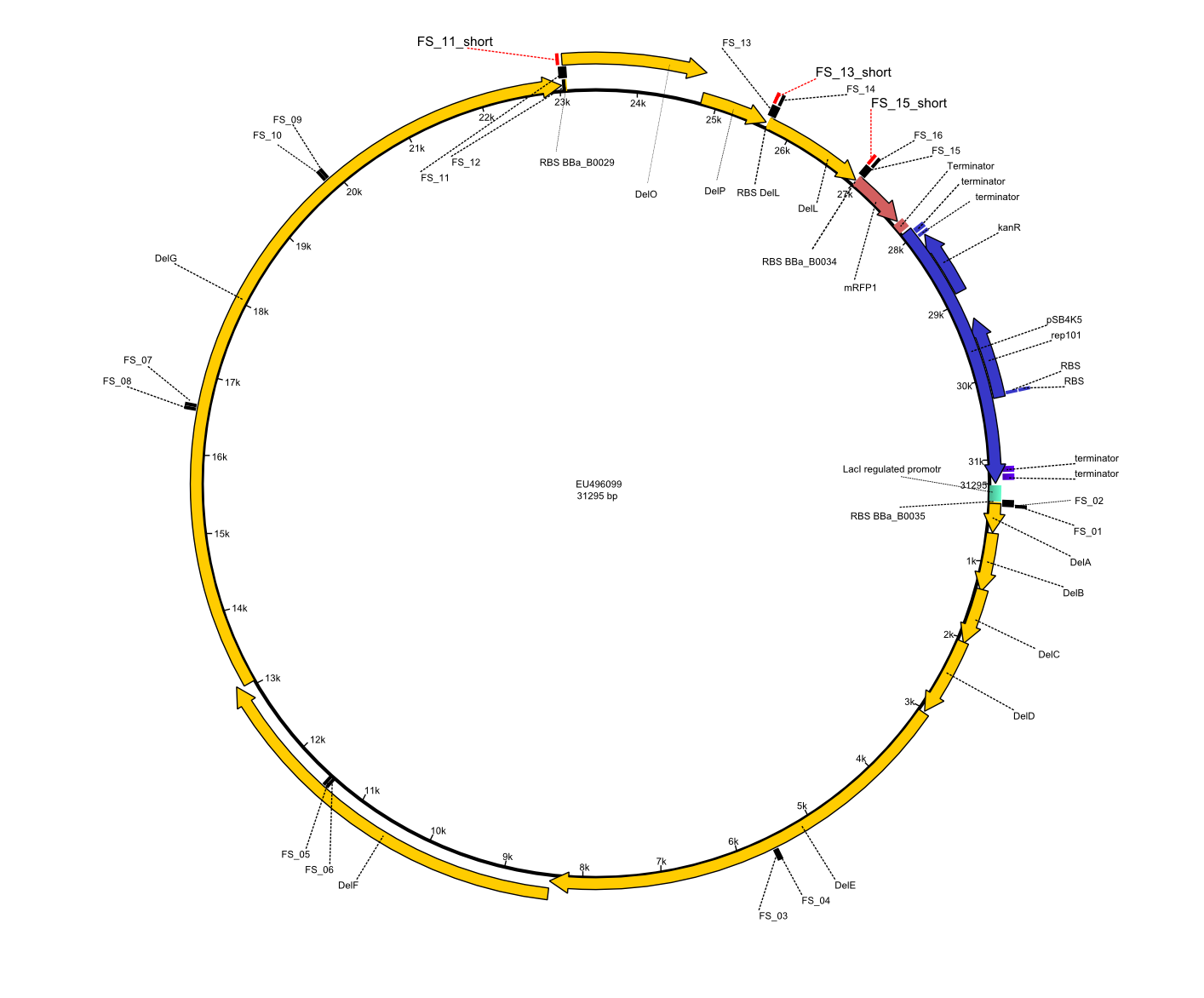
We had various problems with the amplification of the fragments located in theregion DelF-G and DelO-P. For these particular PCRs long primers containing the Gibson overlaps had to be used. As these long primers often form complex secondary structures decreasing annealing efficiency we designed short version of these primers, having the same sequence region complementary to the genomic target, but lacking any longer overhangs. Consequently these primesrs will serve only for amplification of the desired genes from the genome of D. Acidovorans to obtain specific templates. This specific template afterwards will be used to introduce the designated overlap using the long primers again. In total 3 short primer versions FS_11s, FS_13s and FS_15s were ordered.
Primer pairs, corresponding sequences and usage
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| FS_11: DelAG_5_short_rv | 2013-05-07 | Amplification of DelAG from Delftia acidovorans. Gibson Primer | TCAGATATCTCCCAGAGTTTCGAGAAAG |
| FS_13: DelOP_short_rv | 2013-05-07 | Amplification of DelOP from Delftia acidovorans. Gibson Primer with overhang to DelL | TCACACCGTGGTGTCTGCAGGCG |
| FS_15: DelL_mRFP_pSB4K5__short_rv | 2013-05-07 | Amplification of DelL. Gibson Primer with overhang to BBa_J04450 | TCAGTCCTGCAGCGCCAGCTGTTCTGTG |
Amplification of Del Genes from DSM-39 genome
Goals
The PCRs which worked in the previous week will be repeated to obtain higher product yields, as high concentrations of the long amplicons are needed for a successfull Gibson Assembly.
Furthermore we will try to establish PCRs for the obtained short Primers and thereby provide specific templates for the second amplification step using the corresponding primers introducing the required Gibson overlaps.
Amplifications of the sequence region DelF-G will be tried with different protocols to optimize PCR conditions, as we did not obtain any amplicons at all last week. Therefore it is likely that these primers are only able to bind in a very small frame of annealing temperature or even that the used primer combination do not posses a common annealing temperature and therefore amplification with the desired combinations is not possible.
As the amplification of DelO-P also turned out to be difficult, we decided to try a backup strategy which not only includes the desired genes DelO and DelP but also DelL and, since they are located between DelP and DelL, DelM and DelN. These two genes would increase our amplicon size by only 2.2 kb, but we hoped that on the other hand the corresponding primers would work better.
Results
| PCRs from D.acidovorans DSM-39 | ||||
|---|---|---|---|---|
| Gene(s) | Fragment | Primer combination | Successful? | |
| DelA DelB DelC DelD DelE DelF DelG | DelA-E | FS_02 and FS_03 | ||
| DelA-G | FS_02 and FS_07 | |||
| DelE-G | FS_04 and FS_07 | |||
| FS_04 and FS_11s | ||||
| DelF-G | FS_06 and FS_07 | |||
| FS_06 and FS_09 | ||||
| FS_06 and FS_11 | ||||
| FS_06 and FS_11s | ||||
| DelG | FS_08 and FS_11s | |||
| FS_10 and FS_11 | ||||
| FS_10 and FS_11s | ||||
| DelO DelP DelL | DelO-P | FS_12 and FS_13 | ||
| FS_12 and FS_13s | ||||
| DelL | FS_14 and FS_15 | |||
| DelL-P | FS_13 and FS_15s | |||
| pSB4K5 | pSB4K5 | FS_01 and FS_16 | ||
| Re-PCRs | ||||
|---|---|---|---|---|
| Gene(s) | Fragment | Primer combination | Successful? | |
| DelF DelG | DelF-G | Primer FS_06 and FS_09 | ||
| DelO DelP DelL | DelL-P | Primer FS_13_short and FS_15_short | ||
 "
"