Team:Heidelberg/Templates/Del week16 overview
From 2013.igem.org
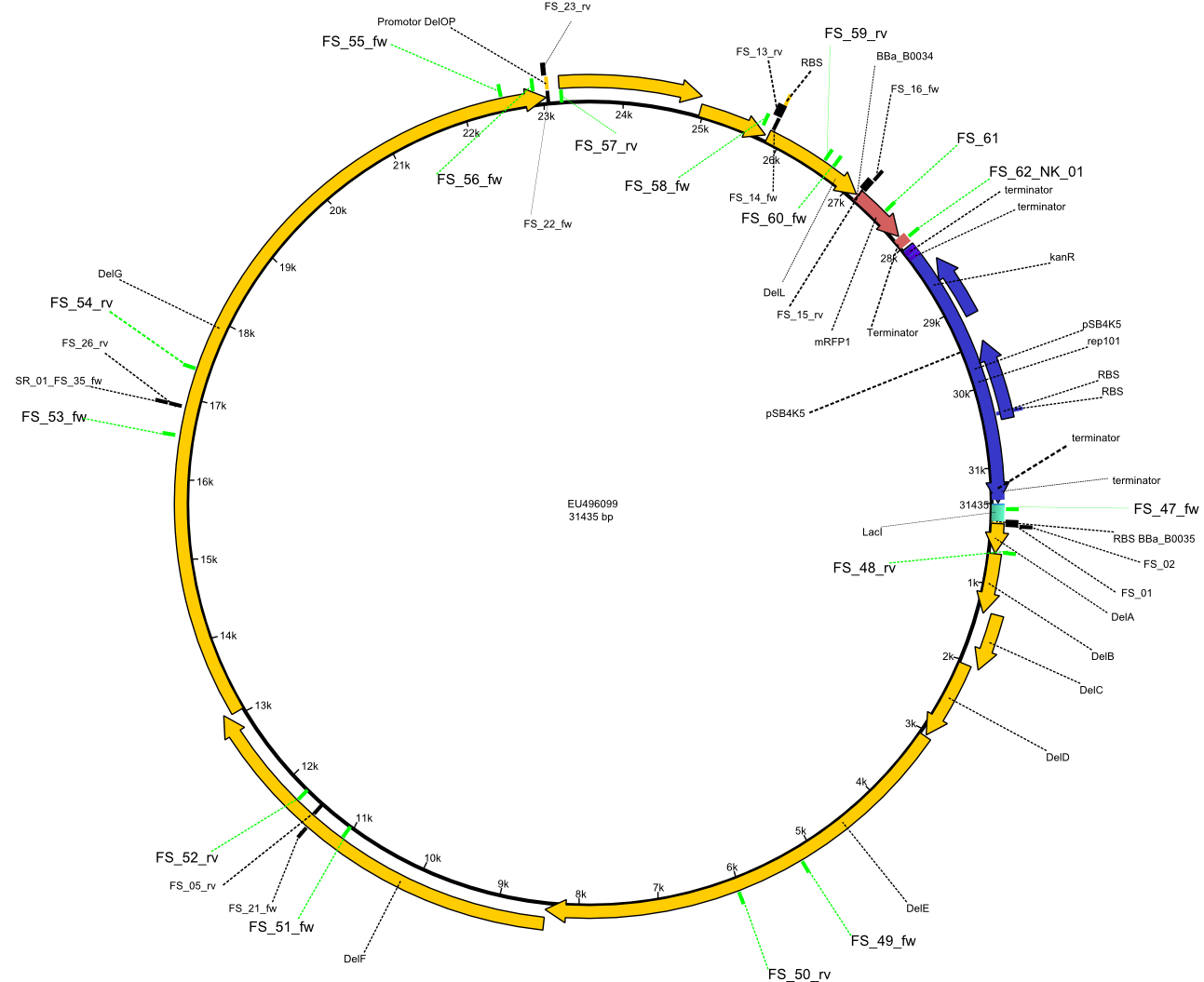
(Created page with " ==Strategy== [[File:Heidelberg_Strategie6.png|500px|right|thumb|Vector map of including Primers FS_01 to FS_16 used for assembly of the desired genes from ''D. acidovorans'' ...") |
|||
| (2 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
==Strategy== | ==Strategy== | ||
| - | + | [[File:Heidelberg_Strategie6.png|300px|right|thumb|Vector map of including Primers FS_01 to FS_16 used for assembly of the desired genes from ''D. acidovorans'' and the partsregistry backbone pSB4K5 .]] | |
| - | [[File:Heidelberg_Strategie6.png| | + | |
Most PCRs have worked!! Yeah- finally!!! Thus, all the fragments needed for Gibson assembly are now available, however yields are still suboptimal for some of the fragments amplified. Furthermore, we designed screening primers for our final construct pFSN. These primers will be used to analyze our transformed bacteria for presence of the complete DelRest construct pFSN by colony-PCR. | Most PCRs have worked!! Yeah- finally!!! Thus, all the fragments needed for Gibson assembly are now available, however yields are still suboptimal for some of the fragments amplified. Furthermore, we designed screening primers for our final construct pFSN. These primers will be used to analyze our transformed bacteria for presence of the complete DelRest construct pFSN by colony-PCR. | ||
| Line 11: | Line 10: | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| + | ====Primer pairs, corresponding sequences and usage==== | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 50: | Line 50: | ||
==Amplification and purification of Del Genes from SPH-1== | ==Amplification and purification of Del Genes from SPH-1== | ||
| - | ===Goals=== | + | ====Goals==== |
As we were able to establish amplification protocols for most of our desired amplicons by the end of last week, we will repeat these PCRs to obtain the required amounts and concentrations. Furthermore we will optimize the PCR conditions of DelO-P by using higher annealing temperatures, hopefully inhibiting secondary structe formation of the primers. Afterwards we will assemble our construct, transform it into '' E.Coli'' DH10B and screen for (hopefully many) positive clones using the recently ordered screening primers. | As we were able to establish amplification protocols for most of our desired amplicons by the end of last week, we will repeat these PCRs to obtain the required amounts and concentrations. Furthermore we will optimize the PCR conditions of DelO-P by using higher annealing temperatures, hopefully inhibiting secondary structe formation of the primers. Afterwards we will assemble our construct, transform it into '' E.Coli'' DH10B and screen for (hopefully many) positive clones using the recently ordered screening primers. | ||
| + | |||
| + | ====Results==== | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 75: | Line 77: | ||
| Primer FS_22 and FS_13 || [[File:Heidelberg_Yes check.svg.png|20px|center]] | | Primer FS_22 and FS_13 || [[File:Heidelberg_Yes check.svg.png|20px|center]] | ||
|} | |} | ||
| - | + | <br/> | |
| + | <br/> | ||
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
Latest revision as of 18:38, 4 October 2013
Contents |
Strategy
Most PCRs have worked!! Yeah- finally!!! Thus, all the fragments needed for Gibson assembly are now available, however yields are still suboptimal for some of the fragments amplified. Furthermore, we designed screening primers for our final construct pFSN. These primers will be used to analyze our transformed bacteria for presence of the complete DelRest construct pFSN by colony-PCR.
Primer pairs, corresponding sequences and usage
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| FS_47_screening_BB_AF_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTTGGCCGATTCATTAATGC |
| FS_48_screening_BB_AF_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | TAACGGTATCGGTATCGCTTTG |
| FS_49_screening_AFI_AFII_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTTTCTCTGGAAGATGGATAC |
| FS_50_screening_AFI_AFII_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTTGACGAAAAAGCCGACCAC |
| FS_51_screening_AF_FG(21-26)_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | TGGATATCGACTGGACTGCCTG |
| FS_52_screening_AF_FG(21-26)_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | TGCACCACATCGACGAAACGG |
| FS_53_screening_FG(21-26)_G_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTACGGCCTATCACATCAGCG |
| FS_54_screening_FG(21-26)_G_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GAACCTGGGTGTTCACGAAAAAGCC |
| FS_55_screening_G_OP_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTATCTCTACATGCATCGCTAC |
| FS_56_screening_G_OP_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | AGGACATTTTCCGCACCCCG |
| FS_57_screening_G_OP_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GCTGGCGTTTTCCATAAG |
| FS_58_screening_OP_L_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GAACAACTTCCAGCACAGCCTGTTC |
| FS_59_screening_OP_L_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | CGTTGAAGATTTCGTTGACG |
| FS_60_screening_L_BB_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | CATCTTCAAGGTGTTCTATGAAC |
| FS_61_screening_L_BB(with_mRFP)_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | CAGTTTAACTTTGTAGATGAAC |
| NK_01_FS_62_screening_L_BB(without_mRFP)_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTTCACCGACAAACAACAGATAAAACG |
Amplification and purification of Del Genes from SPH-1
Goals
As we were able to establish amplification protocols for most of our desired amplicons by the end of last week, we will repeat these PCRs to obtain the required amounts and concentrations. Furthermore we will optimize the PCR conditions of DelO-P by using higher annealing temperatures, hopefully inhibiting secondary structe formation of the primers. Afterwards we will assemble our construct, transform it into E.Coli DH10B and screen for (hopefully many) positive clones using the recently ordered screening primers.
Results
| PCRs from D.acidovorans SPH-1 | ||||
|---|---|---|---|---|
| Gene(s) | Fragment | Primer combination | Successful? | |
| DelA DelB DelC DelD DelE DelF DelG | DelA-F | Primer FS_02 and FS_05 | ||
| DelA-G | Primer FS_02 and FS_11 | |||
| Primer FS_02 and FS_11_short | ||||
| DelF-G | Primer FS_21 and FS_07 | |||
| Primer FS_21 and FS_26 | ||||
| DelG | Primer SR_01 and FS_23 | |||
| DelO DelP DelL | DelO-P | Primer FS_22 and FS_13_short | ||
| Primer FS_22 and FS_13 | ||||
| Test restriction digests of PCR amplified fragments | ||||
|---|---|---|---|---|
| Fragment | Primer | Digestion enzyme | Expected bands | |
| DelO-P | Primer FS_22 and FS_13 | EcoRI-HF | 1883, 960 | |
 "
"