Team:Newcastle/Collaborations
From 2013.igem.org
| (100 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
= Collaborations = | = Collaborations = | ||
| - | + | During our project we were able to collaborate with other iGEMers. This page details the help we gave to other teams. If you would like to see how we contributed to other teams please click on the appropriate University logo's. | |
| - | + | ||
| - | == | + | ==<div id="title" style=" display: none;">Edinburgh</div>== |
| + | <html> | ||
| + | <a href="https://2013.igem.org/Team:Edinburgh/Collaboration"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/a/ae/BareCillus_Edinburgh_logo.jpg" alt="Edinburgh University"> | ||
| + | </a> | ||
| + | </html> | ||
| - | + | We were able to help Team Edinburgh with their modelling by supplying them with some ''Bacillus subtilis'' parameters: | |
| - | == | + | <table class="data"> |
| + | <tr> | ||
| + | <th>Definition</th> | ||
| + | <th>Value</th> | ||
| + | <th>Source</th> | ||
| + | </tr> | ||
| + | <tr class="odd"> | ||
| + | <td class="data">Cell division rate, growth rate</td> | ||
| + | <td class="data">30-73mins</td> | ||
| + | <td class="data">http://www.ncbi.nlm.nih.gov/pmc/articles/PMC106920/pdf/jb000547.pdf</td> | ||
| - | + | </tr> | |
| + | <tr class="even"> | ||
| + | <td class="data">Genome equivalents of DNA per cell</td> | ||
| + | <td class="data">1.6 - 3.3</td> | ||
| + | <td class="data">http://www.ncbi.nlm.nih.gov/pmc/articles/PMC106920/pdf/jb000547.pdf</td> | ||
| + | </tr> | ||
| + | <tr class="odd"> | ||
| + | <td class="data">Replication time of chromosome, "C period"</td> | ||
| + | <td class="data">50-58</td> | ||
| + | <td class="data">http://www.ncbi.nlm.nih.gov/pmc/articles/PMC106920/pdf/jb000547.pdf</td> | ||
| + | </tr> | ||
| + | <tr class="even"> | ||
| + | <td class="data">Delay before cell division, "D period"</td> | ||
| + | <td class="data">21-34</td> | ||
| + | <td class="data">http://www.ncbi.nlm.nih.gov/pmc/articles/PMC106920/pdf/jb000547.pdf</td> | ||
| + | </tr> | ||
| + | <tr class="odd"> | ||
| + | <td class="data">Number of replicating forks</td> | ||
| + | <td class="data">2</td> | ||
| + | <td class="data">-</td> | ||
| + | </tr> | ||
| + | <tr class="even"> | ||
| + | <td class="data">Number of replicating origins</td> | ||
| + | <td class="data">0.43-1.7</td> | ||
| + | <td class="data">http://www.ncbi.nlm.nih.gov/pmc/articles/PMC106920/pdf/jb000547.pdf</td> | ||
| + | </tr> | ||
| + | <tr class="odd"> | ||
| + | <td class="data">Cell volume</td> | ||
| + | <td class="data">0.85±0.38 μm^3 </td> | ||
| + | <td class="data">http://www.ncbi.nlm.nih.gov/pubmed/21395229</td> | ||
| + | </tr> | ||
| + | <tr class="even"> | ||
| + | <td class="data">Minimal replication time of chromosome</td> | ||
| + | <td class="data">55min</td> | ||
| + | <td class="data">http://www.ncbi.nlm.nih.gov/pmc/articles/PMC106920/pdf/jb000547.pdf</td> | ||
| + | </tr> | ||
| + | <tr class="odd"> | ||
| + | <td class="data">Numbe of rrn operons per chomosome</td> | ||
| + | <td class="data">10 operons</td> | ||
| + | <td class="data">http://www.ncbi.nlm.nih.gov/pubmed/12644506</td> | ||
| + | </tr> | ||
| + | |||
| + | </table> | ||
| + | ==<div id="title" style=" display: none;">Leeds</div>== | ||
| + | <html> | ||
| + | <a href="https://2013.igem.org/Team:Leeds/Attributions"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/0/0d/BareCillus_Leeds_logo.jpg" alt="Leeds University" width= "40%"> | ||
| + | </a> | ||
| + | </html> | ||
| + | On Friday the 12th of July our team was at UCL for the [https://2013.igem.org/Team:Newcastle/Outreach/uk_meet iGEM U.K. meet up]. We delivered a [https://2013.igem.org/Team:Newcastle/Outreach/Workshop workshop about Mathematical modelling], including teaching the basics of using the rule-based modelling language BioNetGen. We also supplied attendees with all the software needed to produce BioNetGen models on a handy USB. This workshop introduced the Leeds iGEM team to BioNetGen, [https://2013.igem.org/Team:Leeds/Attributions who went on to use BioNetGen to model the Cpx pathway in their project]. | ||
| + | These are the slides from our modelling presentation: | ||
| + | <html> | ||
| + | <div style="text-align:center;"> | ||
| + | <iframe src="https://skydrive.live.com/embed?cid=A67E48B91B122E55&resid=A67E48B91B122E55%21112&authkey=AKt313lzFLi-ZsQ&em=2" width="402" height="327" frameborder="0" scrolling="no"></iframe> | ||
| + | <p style="text-align:center;">A downloadable version can be found <a href="https://static.igem.org/mediawiki/2013/9/96/BareCillus_Workshop_Presentation.pdf">here</a>.</p> | ||
| + | </div> | ||
| + | </html> | ||
| + | This is the handout from our modelling workshop: | ||
| + | <html> | ||
| + | <div style="text-align:center;"> | ||
| + | <iframe src="https://skydrive.live.com/embed?cid=7825331CA7243819&resid=7825331CA7243819%21110&authkey=AFI9n46tTkAQ2ts&em=2" width="476" height="288" frameborder="0" scrolling="no"></iframe> | ||
| + | <p style="text-align:center"> | ||
| + | A downloadable version can be found <a href="https://static.igem.org/mediawiki/2013/b/ba/Workshop_Handout.pdf">here</a>. | ||
| + | </p> | ||
| + | </div> | ||
| + | </html> | ||
| + | ==<div id="title" style=" display: none;">Manchester</div>== | ||
| + | <html> | ||
| + | <style type="text/css"> | ||
| + | #video { | ||
| + | margin-left:10px; | ||
| + | } | ||
| + | </style> | ||
| + | <a href="https://2013.igem.org/Team:Manchester/Collaboration"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/4/40/BareCillus_Manchester_logo.jpg" alt="Manchester University" width= "40%"></a> | ||
| + | <p>We have helped the Manchester team with their 'introduction to modelling', giving ideas on how this could be achieved and producing a tutorial video giving a brief introduction to modelling using BioNetGen.</p> | ||
| + | <div id="video"> | ||
| + | <video src="https://static.igem.org/mediawiki/2013/4/47/BioNetGen_Tutorial.mp4" width="480" height="260" controls preload></video> | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | ==<div id="title" style=" display: none;">NRP-UEA-Norwich</div>== | ||
| + | <html> | ||
| + | <a href="https://2013.igem.org/Team:NRP-UEA-Norwich/HumanPractice/SoilSamples/31-45"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/e/e9/BareCillus_UEA_logo.png"width= "200px" height="100px"> | ||
| + | </a> | ||
| + | </html> | ||
| + | |||
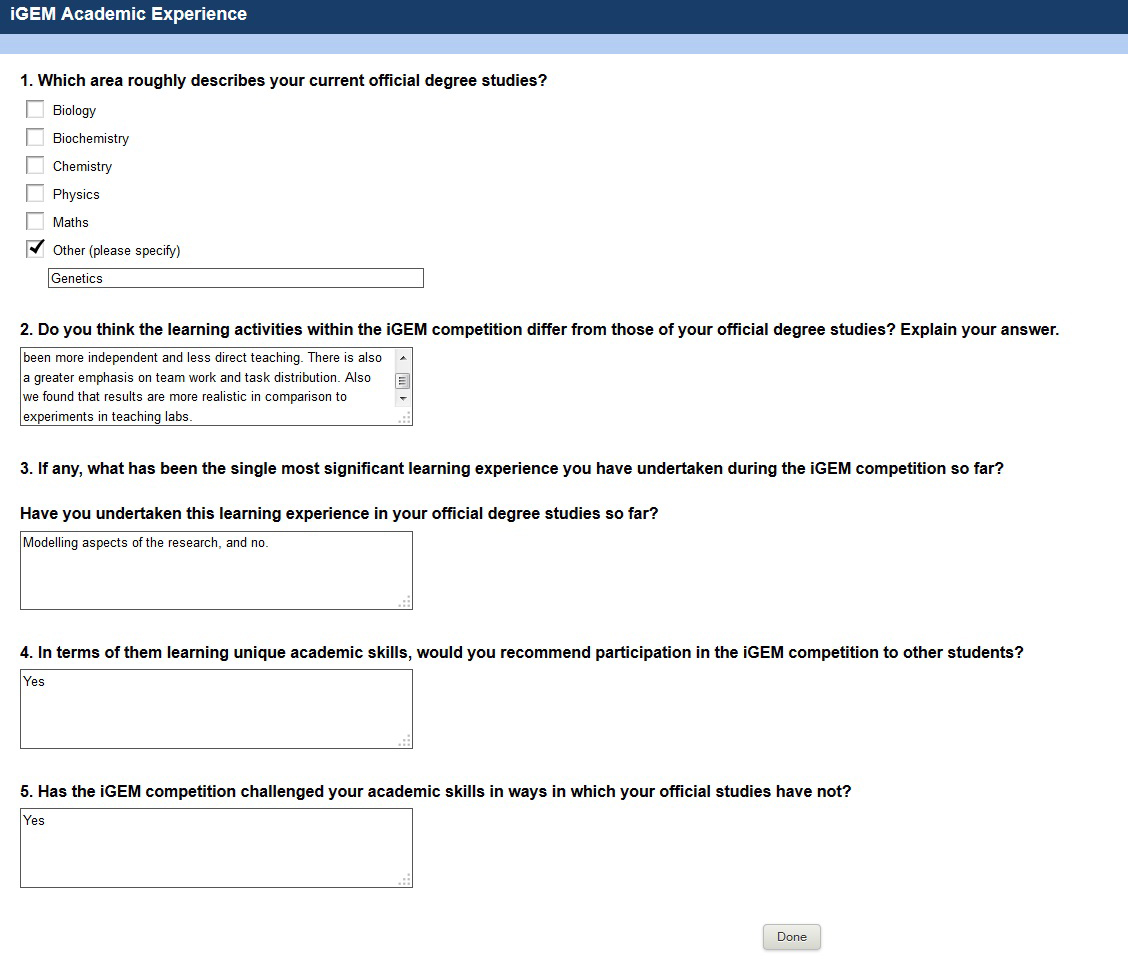
| + | We supplied a [https://2013.igem.org/Team:NRP-UEA-Norwich/HumanPractice/SoilSamples/31-45 soil sample] for the Norwich team to be used in their project. As a part of their Outreach activities, we filled in a questionaire about the differences between the iGEM experience and degree subject learning experince. | ||
| + | |||
| + | [[File:BareCillus_Norwich_1.jpg|600px]] | ||
| + | |||
| + | ==<div id="title" style=" display: none;">Greensboro-Austin</div>== | ||
| + | <html> | ||
| + | <a href="https://2013.igem.org/Team:Greensboro-Austin"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/d/d9/BareCillus_University_of_Texas_at_Austin_logo.png" alt="Texas at Austin"width= "40%"> | ||
| + | </a> | ||
| + | </html> | ||
| + | |||
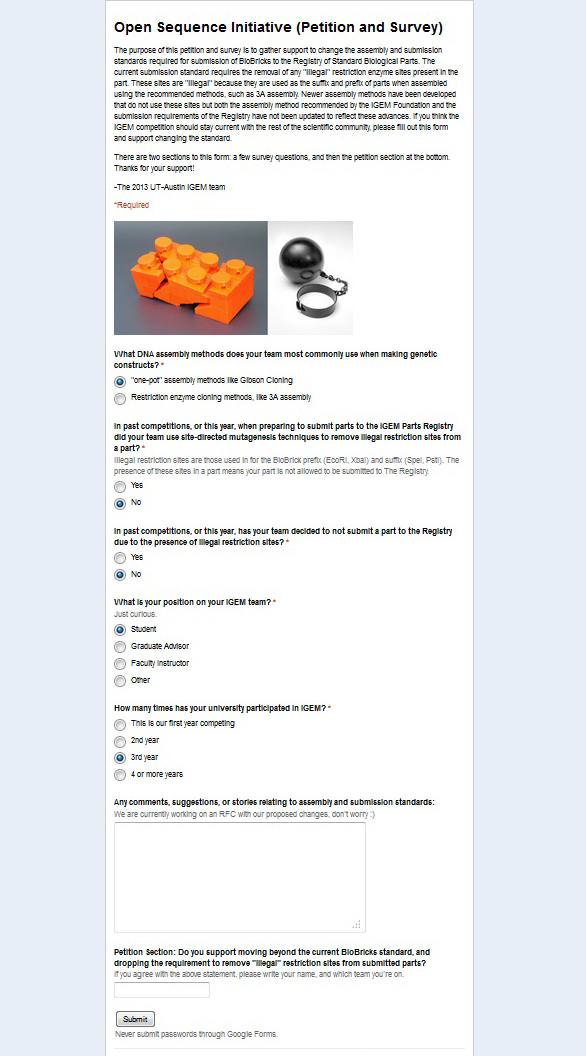
| + | Our team filled in a questionaire about the current BioBrick assembly standards and submission, in order to help the Texas at Austin team modernise the standards. | ||
| + | |||
| + | [[File:BareCillus_Austin_1.jpg]] | ||
{{Team:Newcastle/Sponsors}} | {{Team:Newcastle/Sponsors}} | ||
Latest revision as of 20:35, 4 October 2013

Contents |
Collaborations
During our project we were able to collaborate with other iGEMers. This page details the help we gave to other teams. If you would like to see how we contributed to other teams please click on the appropriate University logo's.
We were able to help Team Edinburgh with their modelling by supplying them with some Bacillus subtilis parameters:
| Definition | Value | Source |
|---|---|---|
| Cell division rate, growth rate | 30-73mins | http://www.ncbi.nlm.nih.gov/pmc/articles/PMC106920/pdf/jb000547.pdf |
| Genome equivalents of DNA per cell | 1.6 - 3.3 | http://www.ncbi.nlm.nih.gov/pmc/articles/PMC106920/pdf/jb000547.pdf |
| Replication time of chromosome, "C period" | 50-58 | http://www.ncbi.nlm.nih.gov/pmc/articles/PMC106920/pdf/jb000547.pdf |
| Delay before cell division, "D period" | 21-34 | http://www.ncbi.nlm.nih.gov/pmc/articles/PMC106920/pdf/jb000547.pdf |
| Number of replicating forks | 2 | - |
| Number of replicating origins | 0.43-1.7 | http://www.ncbi.nlm.nih.gov/pmc/articles/PMC106920/pdf/jb000547.pdf |
| Cell volume | 0.85±0.38 μm^3 | http://www.ncbi.nlm.nih.gov/pubmed/21395229 |
| Minimal replication time of chromosome | 55min | http://www.ncbi.nlm.nih.gov/pmc/articles/PMC106920/pdf/jb000547.pdf |
| Numbe of rrn operons per chomosome | 10 operons | http://www.ncbi.nlm.nih.gov/pubmed/12644506 |
On Friday the 12th of July our team was at UCL for the iGEM U.K. meet up. We delivered a workshop about Mathematical modelling, including teaching the basics of using the rule-based modelling language BioNetGen. We also supplied attendees with all the software needed to produce BioNetGen models on a handy USB. This workshop introduced the Leeds iGEM team to BioNetGen, who went on to use BioNetGen to model the Cpx pathway in their project.
These are the slides from our modelling presentation:
A downloadable version can be found here.
This is the handout from our modelling workshop:
A downloadable version can be found here.
We have helped the Manchester team with their 'introduction to modelling', giving ideas on how this could be achieved and producing a tutorial video giving a brief introduction to modelling using BioNetGen.
We supplied a soil sample for the Norwich team to be used in their project. As a part of their Outreach activities, we filled in a questionaire about the differences between the iGEM experience and degree subject learning experince.
Our team filled in a questionaire about the current BioBrick assembly standards and submission, in order to help the Texas at Austin team modernise the standards.
 "
"