Team:Heidelberg/Templates/MM week17p
From 2013.igem.org
(Difference between revisions)
Nils.kurzawa (Talk | contribs) m (Created page with " == 2013-08-19 == * received BL21(DE3) from Gruss Lab, ZMBH * grow in LB to OD=0.53, prepare competent cells * co-transform with pIK1.2(0.2 µl ...") |
|||
| Line 1: | Line 1: | ||
| - | |||
== 2013-08-19 == | == 2013-08-19 == | ||
* received BL21(DE3) from Gruss Lab, ZMBH | * received BL21(DE3) from Gruss Lab, ZMBH | ||
| Line 13: | Line 12: | ||
== 2013-08-21 == | == 2013-08-21 == | ||
| - | [[File: | + | [[File:Heidelberg_Methylmalonyl-digest2013-08-21.png|150px|thumb|left|Lane 1: NEB 2-log; lanes 2-3: pSB3K3-BBa_J04450 digested with EcoRI+SpeI; lane 4: pIK1.2 digested with EcoRI+SpeI; lane 5: pIK2.6 digested with EcoRI+SpeI]] |
* [[Preparing glycerol stocks|prepare glycerol stock]] of TOP10-pSB3K3-BBa_J04450 | * [[Preparing glycerol stocks|prepare glycerol stock]] of TOP10-pSB3K3-BBa_J04450 | ||
* perform miniPrep -> 10.1 ng/µl in 38 µl | * perform miniPrep -> 10.1 ng/µl in 38 µl | ||
| Line 21: | Line 20: | ||
== 2013-08-22 == | == 2013-08-22 == | ||
| - | [[File: | + | [[File:Heidelberg_Methylmalonyl-digest2013-08-22.png|150px|thumb|right|Lane 1: pSB3K3-BBa_J04450 digested with EcoRI+SpeI, lane 2: NEB 2-log]] |
| - | [[File: | + | [[File:Heidelberg_20130822 pRB19-21 pSB3K3.png|150px|thumb|left|Lane 1: NEB 2-log; lanes 2-9: pRB* plasmids; lane 10: 1 µl of pSB3K3-BBa_J04450 midiPrep]] |
* no colonies of BL21(DE3)-pMM64-pMM65: co-transform BL21(DE3) with pMM64(1 µl = 15 ng)+pMM65(1 µl = 75 ng), plate on Amp+Kan+IPTG | * no colonies of BL21(DE3)-pMM64-pMM65: co-transform BL21(DE3) with pMM64(1 µl = 15 ng)+pMM65(1 µl = 75 ng), plate on Amp+Kan+IPTG | ||
* prepare midiPrep of pSB3K3-BBa_J04450: 2952.1 ng/µl | * prepare midiPrep of pSB3K3-BBa_J04450: 2952.1 ng/µl | ||
| Line 50: | Line 49: | ||
== 2013-08-23 == | == 2013-08-23 == | ||
| - | [[File: | + | [[File:Heidelberg_Methylmalonyl-colony-PCR2013-08-23.png|150px|thumb|left|Colony-PCR of TOP10 transformed with pSB3K3 ligated with pIK1/pIK2 and primers RB43+VR. Lane 1: NEB 2-log; lanes 2-6: pIK1; lanes 7-11: pIK2]] |
| - | [[File: | + | [[File:Heidelberg_Methylmalonyl-colony-PCR2013-08-23_2.png|150px|thumb|left|Colony-PCR of TOP10 transformed with pSB3K3 ligated with pIK2 and primers RB43+VR. Lane 1: NEB 2-log; lanes 2-11: pIK2]] |
* pick 5 colonies each, run colony-PCR with primers RB43+VR (expected: 1kb, using iTaq, 20 µl total volume): | * pick 5 colonies each, run colony-PCR with primers RB43+VR (expected: 1kb, using iTaq, 20 µl total volume): | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 76: | Line 75: | ||
== 2013-08-24 == | == 2013-08-24 == | ||
| - | [[File: | + | [[File:Heidelberg_Methylmalonyl-digest2013-08-24.png|150px|thumb|right|Lane 1: NEB 2-log; lane 3: pSB3K3 digested with EcoRI+SpeI; lane 5: pIK2.6 digested with EcoRI+SpeI]] |
* make miniPrep of pIK2.6: 97 ng/µl in 37.5 µl | * make miniPrep of pIK2.6: 97 ng/µl in 37.5 µl | ||
* digest 3 µg pSB3K3 (1 µl of midiPrep from 2013-08-22), 1.7 µg pIK2.6 (17 µl of miniPrep) with EcoRI+SpeI (20 µl total volume, 0.5 µl of each enzyme) | * digest 3 µg pSB3K3 (1 µl of midiPrep from 2013-08-22), 1.7 µg pIK2.6 (17 µl of miniPrep) with EcoRI+SpeI (20 µl total volume, 0.5 µl of each enzyme) | ||
| Line 98: | Line 97: | ||
== 2013-08-25 == | == 2013-08-25 == | ||
| - | [[File: | + | [[File:Heidelberg_Methylmalonyl-colony-PCR2013-08-24_1.png|150px|thumb|left|Colony-PCR of TOP10 transformed with pSB3K3 ligated with pIK2 and primers RB43+VR. Lane 1: NEB 2-log; lanes 2-11: pIK2]] |
* pick 10 colonies, run colony-PCR with primers RB43+VR (expected: 1kb, using iTaq, 20 µl total volume): | * pick 10 colonies, run colony-PCR with primers RB43+VR (expected: 1kb, using iTaq, 20 µl total volume): | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 119: | Line 118: | ||
* VR primer might not be binding to pSB3K3: http://www.ccbi.cam.ac.uk/iGEM2006/index.php/Primer_List#BioBrick_verification_primers , https://2009.igem.org/Team:Groningen/Notebook/22_July_2009 | * VR primer might not be binding to pSB3K3: http://www.ccbi.cam.ac.uk/iGEM2006/index.php/Primer_List#BioBrick_verification_primers , https://2009.igem.org/Team:Groningen/Notebook/22_July_2009 | ||
* pick another 10 colonies, run colony-PCR with primers VF2+IK25 (expected: 1kb, iTaq, 20 µl total volume): | * pick another 10 colonies, run colony-PCR with primers VF2+IK25 (expected: 1kb, iTaq, 20 µl total volume): | ||
| - | [[File: | + | [[File:Heidelberg_Methylmalonyl-colony-PCR2013-08-24_2.png|150px|thumb|right|Colony-PCR of TOP10 transformed with pSB3K3 ligated with pIK2 and primers VF2+IK25. Lane 1: NEB 2-log; lanes 2-11: pIK2]] |
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
Revision as of 23:34, 4 October 2013
Contents |
2013-08-19
- received BL21(DE3) from Gruss Lab, ZMBH
- grow in LB to OD=0.53, prepare competent cells
- co-transform with pIK1.2(0.2 µl = 120 ng)+pMM64(1 µl = 15 ng), pIK2.6(0.2 µl = 60 ng)+pMM64(1 µl = 15 ng), plate on Cm+Amp+IPTG, grow at 37°C
2013-08-20
- all colonies white
- pick 1 colony each, inoculate LB+Cm+Amp, grow at 37°C until cultures are opaque, add IPTG to 1mM, grow at 30°C
- no visible indigoidine production
- need to clone into a different backbone in order to be able to cotransform with pRB21
- inoculate LB+Kan with TOP10-pSB3K3-BBa_J04450, grow at 37°C
2013-08-21
- prepare glycerol stock of TOP10-pSB3K3-BBa_J04450
- perform miniPrep -> 10.1 ng/µl in 38 µl
- digest with EcoRI+SpeI: 380 ng of pSB3K3-BBa_J04450: use everything (50 µl total volume, 2x0.5 µl enzyme, expect 2.7kb & 1kb), 1200 ng of pIK1.2 (2 µl of miniPrep from 2013-08-09, 20 µl total volume, 2x0.5 µl enzyme, expect 6.7kb & 2.7kb), 900 ng of pIK2.6 (3 µl of miniPrep from 2013-08-09, 20 µl total volume, 2x0.5 µl enzyme, expect 4.4kb & 2.7kb)
- pSB3K3-BBa_J04450 looks as though it was not cut: inoculate 50 ml TB+Kan with TOP10-pSB3K3-BBa_J04450, grow at 37°C
- as control for indigoidine production from pMM64: co-transform BL21(DE3) with pMM64(1 µl = 15 ng)+pMM65(0.2 µl = 13 ng), plate on Amp+Kan+IPTG
2013-08-22
- no colonies of BL21(DE3)-pMM64-pMM65: co-transform BL21(DE3) with pMM64(1 µl = 15 ng)+pMM65(1 µl = 75 ng), plate on Amp+Kan+IPTG
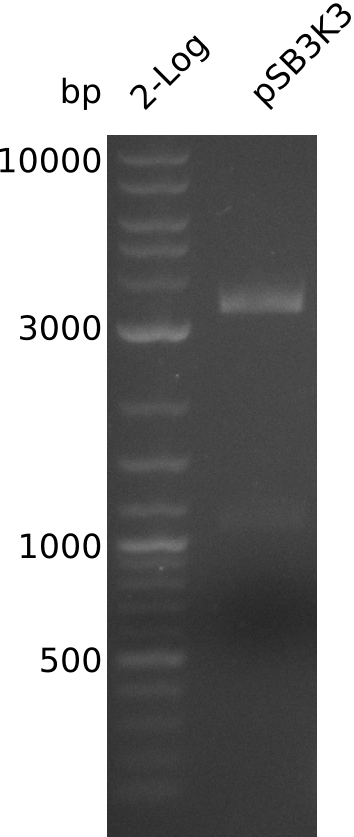
- prepare midiPrep of pSB3K3-BBa_J04450: 2952.1 ng/µl
- digest 0.4 µl of midiPrep with EcoRI+SpeI (20 µl total volume, 2x0.5 µl enzyme)
- band too weak: 2 possibilities:
- concentration measurement wrong
- mispipeted
- => load 1 µl of midiPrep on gel -> concentration measurement about right
- large pSB3K3 fragment over 3 kb, consistent with miniPrep from 2013-08-21 -> 2.7 kb expected, sequence wrong on parts.igem.org
- gel-purify pSB3K3, 6 kb fragment of pIK1, 4 kb fragment of pIK2
- 18 µl of pSB3K3 (not measured), 12.3 ng/µl pIK1 in 21 µl; 7.9 ng/µl pIK2 in 21 µl
- ligate at RT for 1h:
| what | µl |
|---|---|
| pSB3K3 | 9 |
| insert | 8 |
| T4 ligase | 1 µl |
| T4 ligase buffer | 2 µl |
- heat-inactivate: 75°C for 5 min
- transform 10 µl of each ligation into TOP10, plate on Kan, grow at 37°C (pSB3K3+pIK2 = pIK6)
2013-08-23
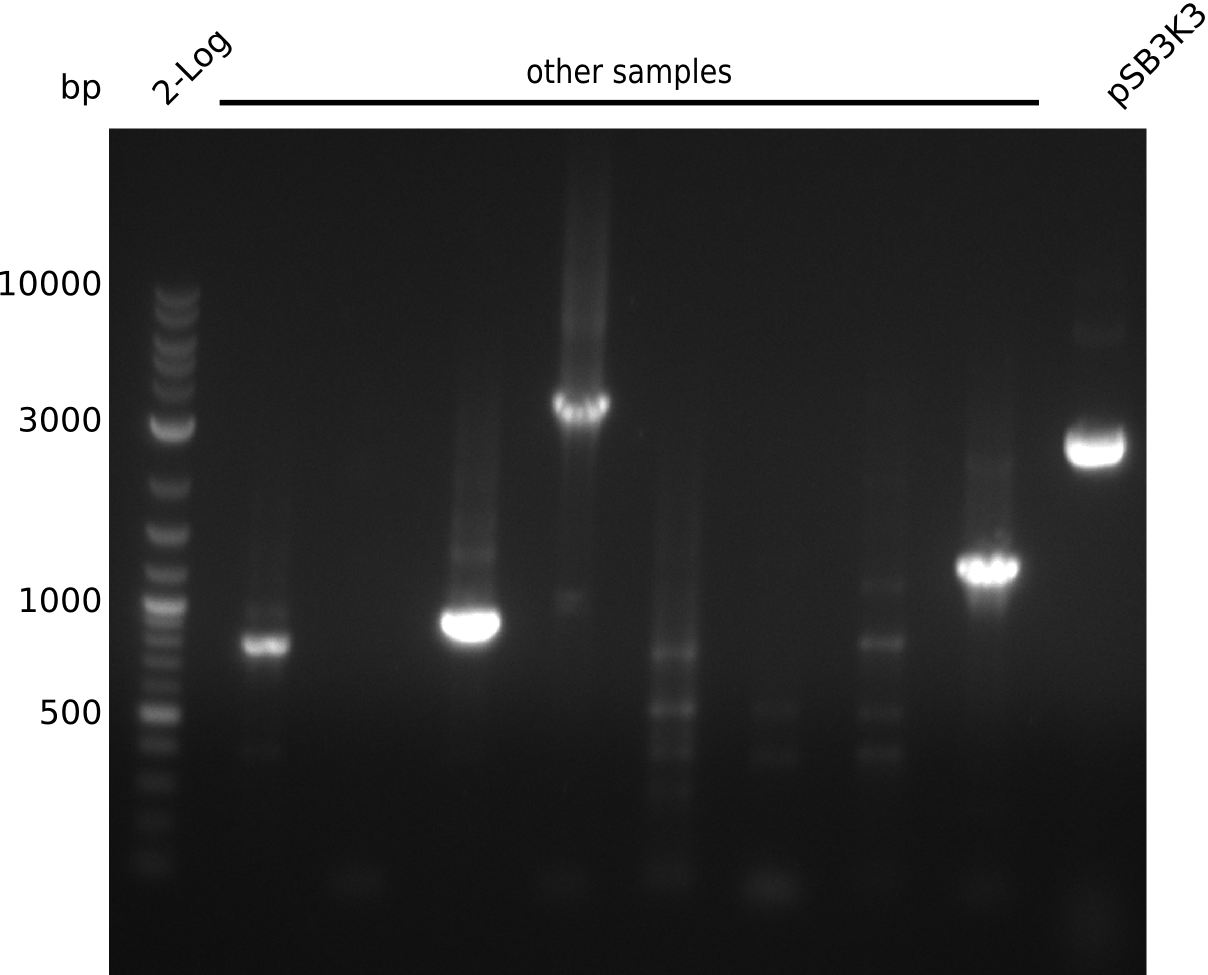
- pick 5 colonies each, run colony-PCR with primers RB43+VR (expected: 1kb, using iTaq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 30 |
| 54 | 30 | |
| 72 | 90 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- no amplificate
- have another look at sequences (motivated by DelRest results): 1 bp missing in pIK1.2: frame shift in permeability device => send pIK1.3 to sequencing with primers VF2, VR, IK25
- pick 30 colonies of pSB3K3-pIK2, run colony-PCR with primers RB43+VR (pool 3 colonies in 1 PCR reaction, iTaq, 20 µl total volume)
- no positives => ligation did not work, most likely due to low DNA concentrations
- need to repeat: inoculate 2x5 ml 2xYT+Cm with TOP10-pIK2.6
2013-08-24
- make miniPrep of pIK2.6: 97 ng/µl in 37.5 µl
- digest 3 µg pSB3K3 (1 µl of midiPrep from 2013-08-22), 1.7 µg pIK2.6 (17 µl of miniPrep) with EcoRI+SpeI (20 µl total volume, 0.5 µl of each enzyme)
- treat pSB3K3 digest with antarctic phosphatase (37°C, 60 min)
- run gel, gel-purify -> 18 µl pSB3K3 (not measured), 15.0 ng/µl pIK2.6 in 20.5 µl
- ligate at RT for 1h:
| what | µl |
|---|---|
| pSB3K3 | 9 |
| pIK2.6 | 8 |
| T4 ligase | 1 µl |
| T4 ligase buffer | 2 µl |
- heat-inactivate: 75°C for 5 min
- transform 10 µl of ligation into TOP10, plate on Kan, grow at 37°C
2013-08-25
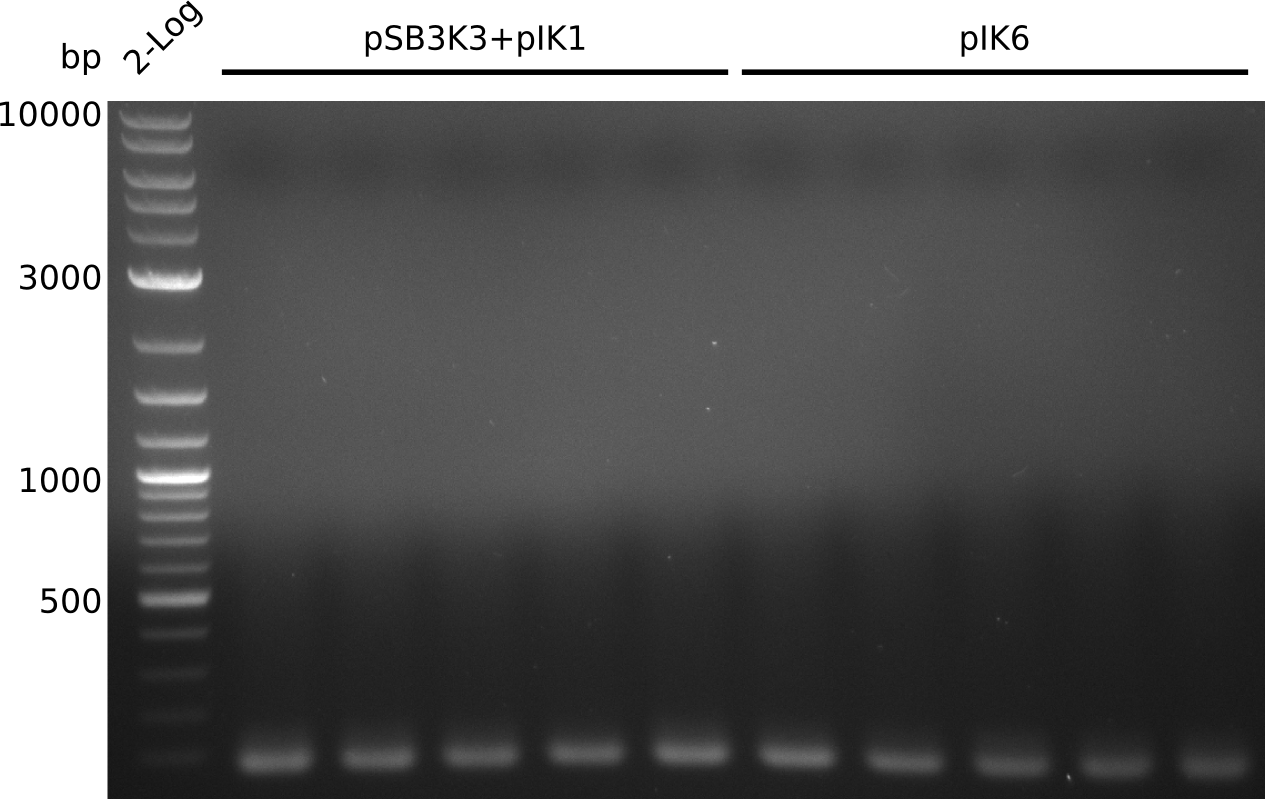
- pick 10 colonies, run colony-PCR with primers RB43+VR (expected: 1kb, using iTaq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 30 |
| 54 | 30 | |
| 72 | 90 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- no amplificate
- VR primer might not be binding to pSB3K3: http://www.ccbi.cam.ac.uk/iGEM2006/index.php/Primer_List#BioBrick_verification_primers , https://2009.igem.org/Team:Groningen/Notebook/22_July_2009
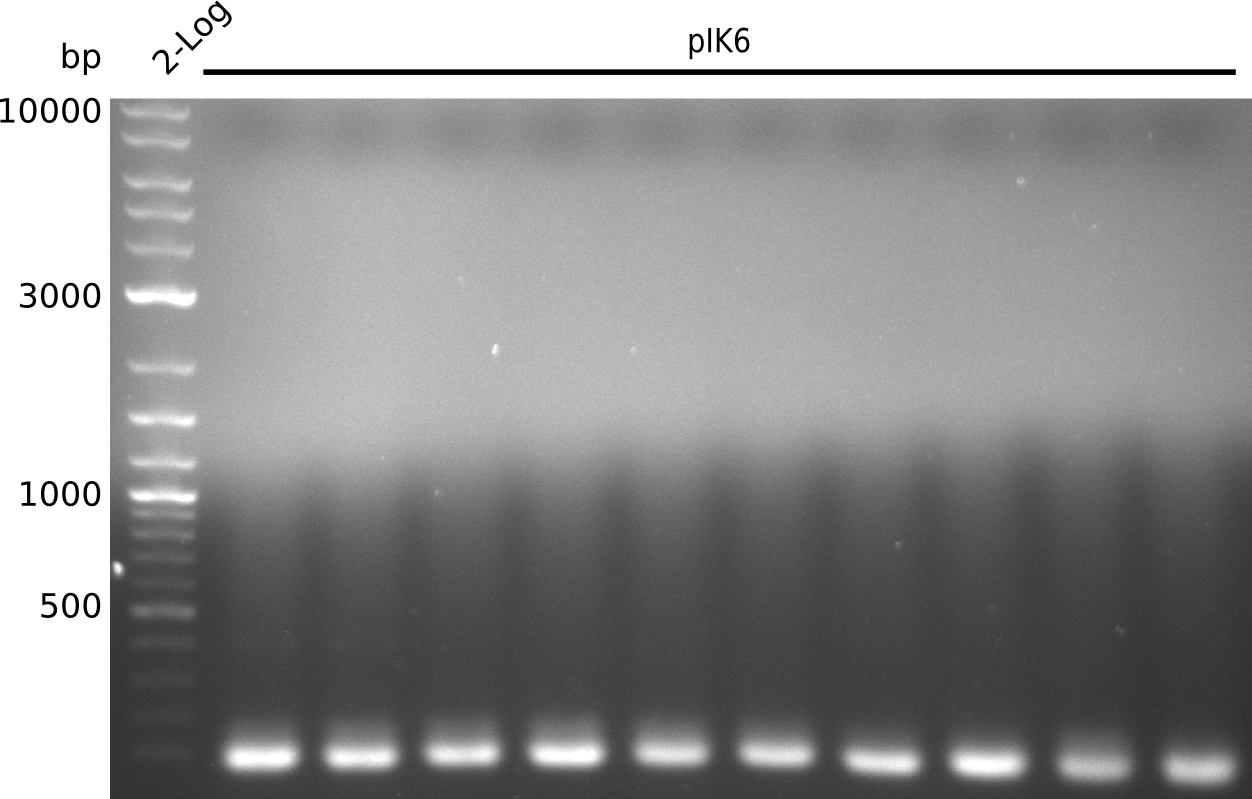
- pick another 10 colonies, run colony-PCR with primers VF2+IK25 (expected: 1kb, iTaq, 20 µl total volume):
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 300 |
| 35 | 95 | 30 |
| 54 | 30 | |
| 72 | 90 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
- definite product, but at 1.3 kb => pSB3K3 was longer than expected in digest, might be a result from that
- unspecific bands probably due to melting temperature of IK25 being at 67°C
- inoculate 2xYT+Kan, grow at 37°C
- pSB3K3 used was from plate 5 => transform TOP10 with pSB3K3 from plate 2, well 6 F
 "
"