Team:Heidelberg/Templates/MM week13p overview
From 2013.igem.org
(Difference between revisions)
| Line 1: | Line 1: | ||
| + | |||
== Goal == | == Goal == | ||
| - | [[File:Heidelberg_PIK1.png|150px|thumb | + | [[File:Heidelberg_PIK1.png|150px|thumb|Vector map of the [[:File:Heidelberg_pIK1.gb|pIK1 plasmid]].]] |
| - | [[File:Heidelberg_PIK2.png|150px|thumb | + | [[File:Heidelberg_PIK2.png|150px|thumb|Vector map of the [[:File:Heidelberg_pIK2.gb|pIK2 plasmid]].]] |
Back-up plan in case the genomic integration does not work. Two plasmids are to be constructed, pIK1 (containing BBa_I746200, pccB-accA, and sfp) and pIK2 (containing pccB-accA and sfp) in the pSB3C5 backbone. 6 fragments are to be amplified: | Back-up plan in case the genomic integration does not work. Two plasmids are to be constructed, pIK1 (containing BBa_I746200, pccB-accA, and sfp) and pIK2 (containing pccB-accA and sfp) in the pSB3C5 backbone. 6 fragments are to be amplified: | ||
{| class="wikitable" | {| class="wikitable" | ||
Revision as of 03:33, 5 October 2013
Goal
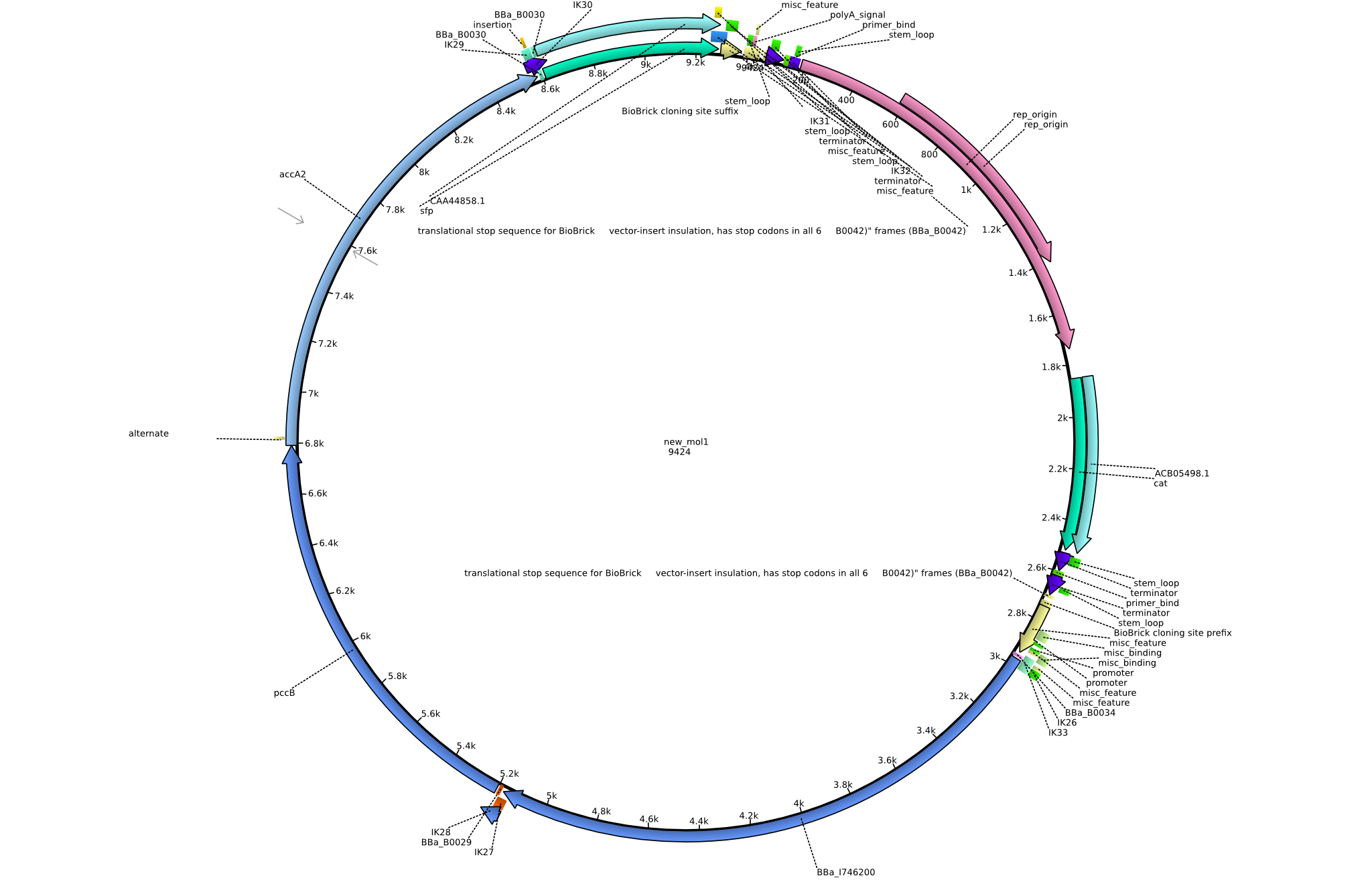
Vector map of the pIK1 plasmid.
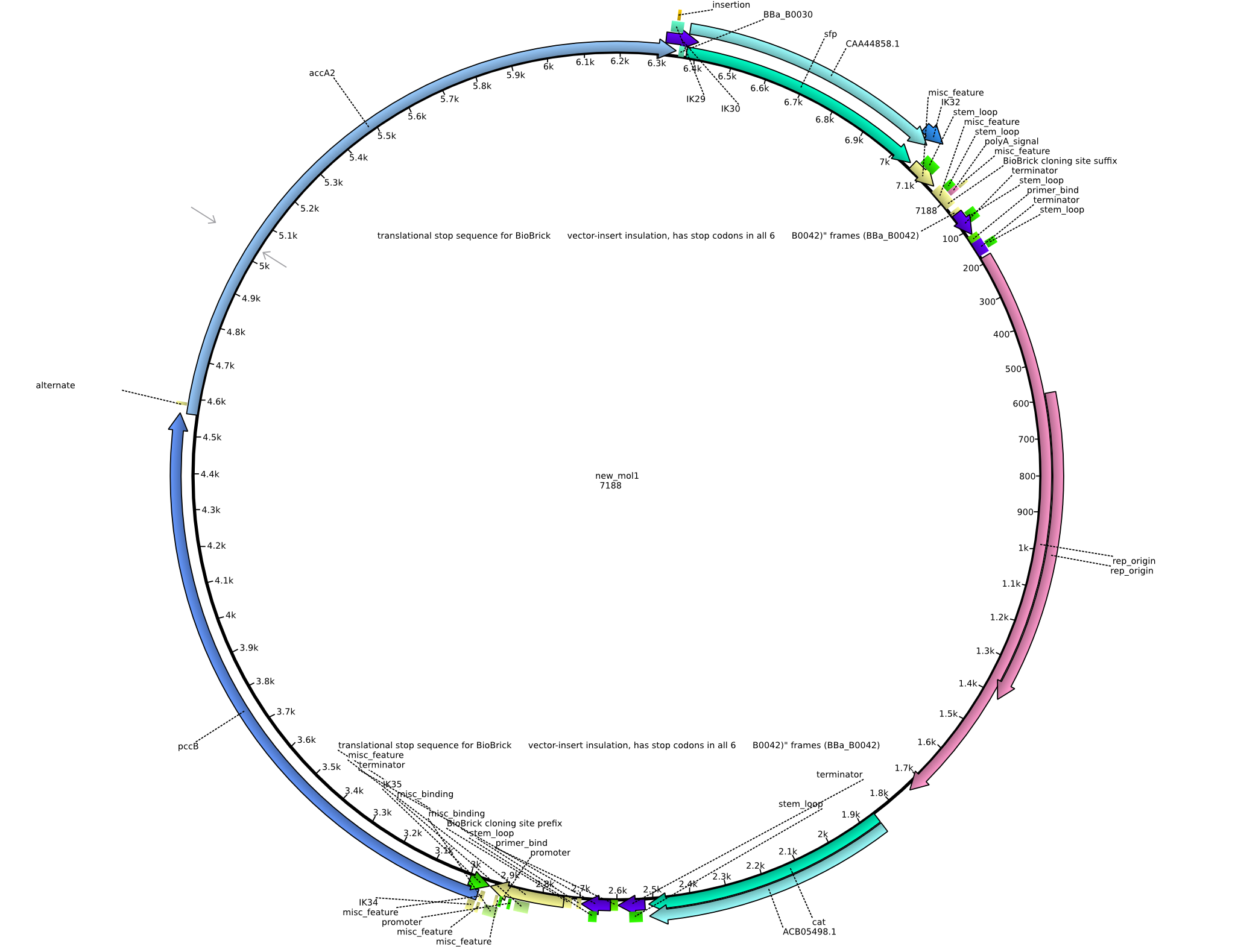
Vector map of the pIK2 plasmid.
Back-up plan in case the genomic integration does not work. Two plasmids are to be constructed, pIK1 (containing BBa_I746200, pccB-accA, and sfp) and pIK2 (containing pccB-accA and sfp) in the pSB3C5 backbone. 6 fragments are to be amplified:
| fragment ID | source | primers | size [kb] |
|---|---|---|---|
| f1 | BBa_J04450 in pSB3C5 | IK32+IK33 | 3 |
| f2 | BBa_I746200 | IK26+IK27 | 2.3 |
| f3 | pccB-accA in pLF03 | IK38+IK29 | 3.4 |
| f4 | sfp in BAP1 | IK30+IK31 | 0.7 |
| f5 | BBa_J04450 in pSB3C5 | IK32+IK35 | 3 |
| f6 | pccB-accA in pLF03 | IK34+IK29 | 3.4 |
 "
"