Team:Heidelberg/Templates/Indigoidine week9
From 2013.igem.org
| Line 1: | Line 1: | ||
| - | |||
We want to assemble pKH1, which is a pSB1C3-derived plasmid containing pSB1C3-lacPromotor-BBa_B0034-bpsA(pMM64)- | We want to assemble pKH1, which is a pSB1C3-derived plasmid containing pSB1C3-lacPromotor-BBa_B0034-bpsA(pMM64)- | ||
| Line 61: | Line 60: | ||
* gel purification | * gel purification | ||
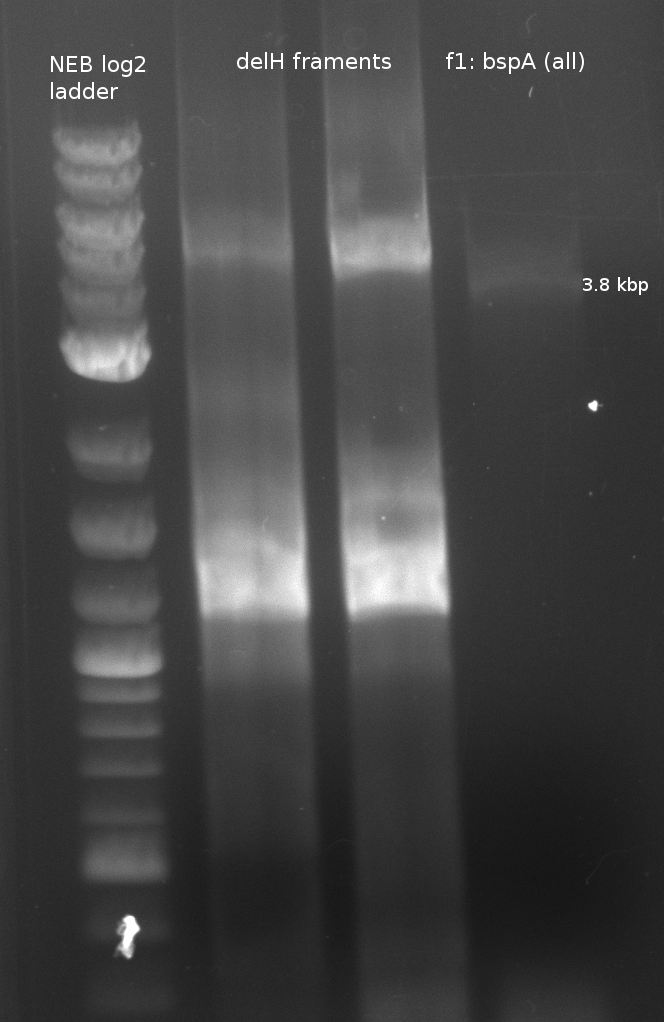
| - | + | [[File:Heidelberg_180613_delH1aA_delH1a_bpsa.png|400px|f1: bpsA (all)]] | |
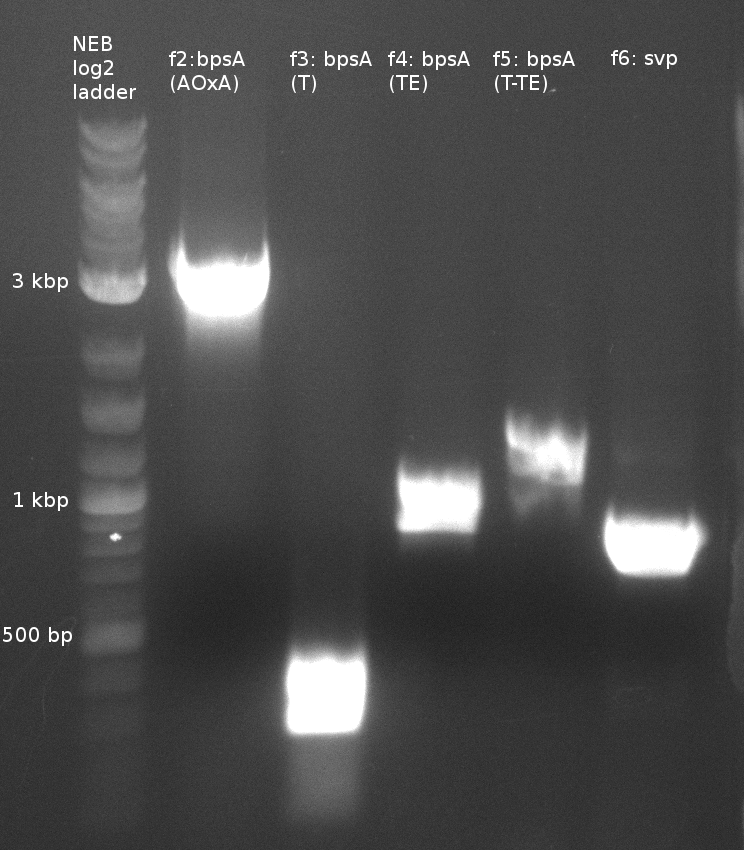
| - | File:Heidelberg_180613_delH1aA_delH1a_bpsa.png|f1: bpsA (all) | + | [[File:Heidelberg_180613_indigoidine-f2-6.png|400px|f2-6: single domains of bpsA and svp]] |
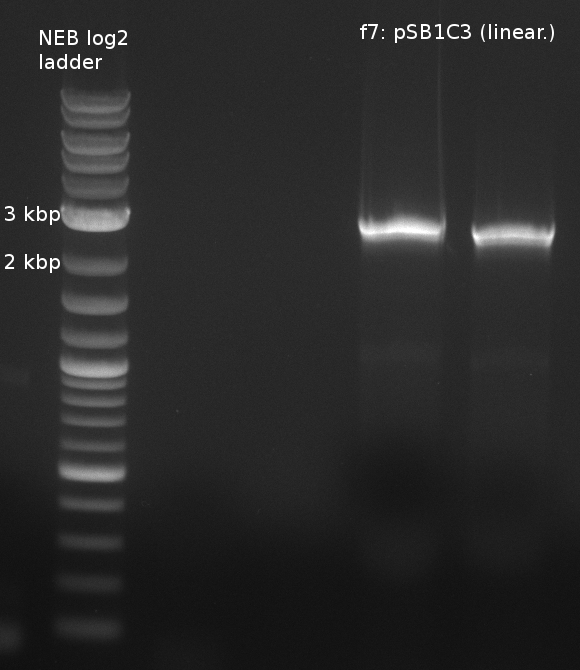
| - | File:Heidelberg_180613_indigoidine-f2-6.png|f2-6: single domains of bpsA and svp | + | [[File:Heidelberg_180613_indigoidine_f7.png|400px|f7: backbone]] |
| - | File:Heidelberg_180613_indigoidine_f7.png|f7: backbone | + | |
| - | + | ||
===Assembly of pKH1 and Transformation (Konrad)=== | ===Assembly of pKH1 and Transformation (Konrad)=== | ||
Latest revision as of 15:35, 21 October 2013
We want to assemble pKH1, which is a pSB1C3-derived plasmid containing pSB1C3-lacPromotor-BBa_B0034-bpsA(pMM64)-
BBa_B0034-svp(pMM65). Single fragments are amplified and cloned using a Gibson approach. We received Streptomyces lavendulae lavendulae since S. lavendulae ATCC11924 has been shown to carry bpsA
[Takahashi 2007].
Fragment amplification for pKH1 (Konrad)
- prepare TB medium for ON (for comparement with LB)
- colony PCR for plated pMM64 and pMM65 strains from 2013-06-16
- prepare combinatorial experiment with gibson cloning or CPEC (polysytronic expression of bpsA and svp in TOP10)
- prepare primer for bpsA only plasmid for BAP1 validation
- 0.4 µl template
- 2x 5 µl Primer (1:10 dilution)
- 25 µl Phusion MM
- 14.6 µl H2O
| fragment | primer | template (DNA) | annealing temp (X1;X2) [°C] | elongation time (Y) [s] |
|---|---|---|---|---|
| f1: bpsA (all) | (NI01,NI06) | pMM64 | 66 (30x, no touch down) | 135 |
| f2: bpsA (AOxA) | (NI01,NI02) | 68;65 | 60 | |
| f3: bpsA (T) | (NI03,NI04) | |||
| f4: bpsA (TE) | (NI05,NI06) | |||
| f5: bpsA (T-TE) | (NI03,NI06) | |||
| f6: svp | (NI07,NI08) | pMM65 | ||
| f7: pSB1C3 (linear.) | (NI09,NI10) | pSB1C3 with J04450 (pJM03) | 66;61 | 90 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 10 | 98 | 1 |
| X1 (incr. down with 0.5 °C) | 5 | |
| 72 | Y | |
| 20 | 98 | 1 |
| X2 | 5 | |
| 72 | Y | |
| 1 | 72 | 240 |
| 1 | 4 | inf |
- gel purification
Assembly of pKH1 and Transformation (Konrad)
Gibson approach
- molecular ratio = backbone : inserts = 1 : 3
- partslength: bpsA=3.8 kbp; svp=0.8 kbp; pSB1C3=2.4 kbp
| fragment | volume [µl] | DNA amount [ng] |
|---|---|---|
| f2: bpsA (AOxA) | 3 | 126 |
| f5: bpsA (T-TE) | 1.5 | 60 |
| f6: svp | 1.5 | 109 |
| f7: pSB1C3 (linear.) | 2 | 51 |
| total | 8 | |
| 2 µl | H2O | |
| 10 µl | Gibson MM |
CPEC approach
Two different approaches were used. In the first approach 3 fragments (backbone (pSB1C3 based), bpsA and svp) were
used for assembly while for the second approach 4 different fragments (Backbone, AOxA, T-TE, svp) were used (like
for the Gibson mix). The amount of used template was calculated for backbone : insert ratio of 1:1 and for insert
fragment ratio at equimolar amount.
- 3 fragment mix
- 3 µl template (f7:backbone)
- 8 µl template (f1:bpsA)
- 0.3 µl template (f6:svp)
- 12.5 µl Phusion MM
- 1.2 µl H2O
- 4 fragment mix
- 3 µl template (f7:backbone)
- 1.7 µl template (f2:AOxA)
- 1.5 µl template (f5:T-TE)
- 0.3 µl template (f6:svp)
- 12.5 µl Phusion MM
- 6 µl H2O
The used cycler program was carried out similar to CPEC paper for multipart assembly but I forgot to note down the
exact parameters. (Shame on me...)
Transformation
- into TOP10
- 4 plates (1:1 and 1:4 of Gibson mix; 1x of each CPEC mix)
Colony PCR
analytical digestion of selected clones
validation of transformed BAP1 with Fussenegger plasmids
fusion PCR
Gibson Assembly of bpsA + svp insert
PCR for colony validation
IPTG induction, titration
IPTG induction, titration RESULT
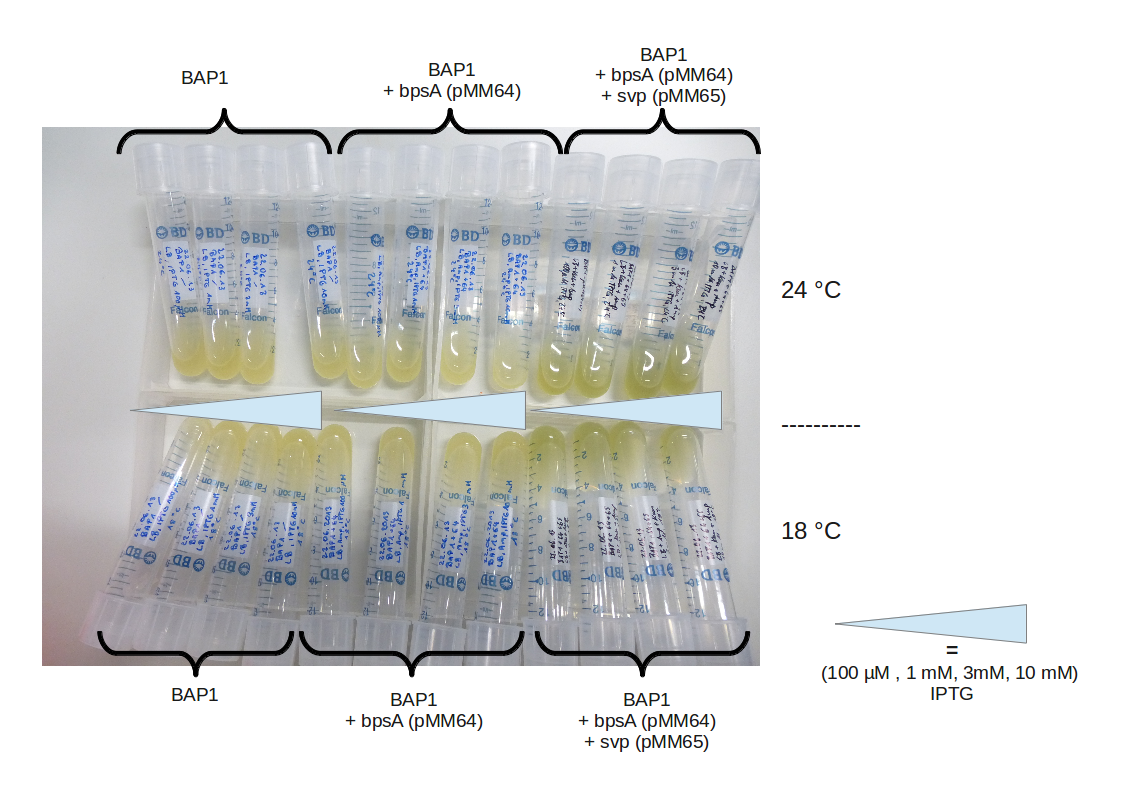
CPEC and transformation
IPTG induction, titration RESULT
Streptomyces cultivation (Ralf)
media for streptomyces culture
- YEME medium (Kieser et al. 2000)
- 10 g Glucose
- 3 g Yeast extract
- 5 g Bacteriological Peptone (I used Pankreatic Digest of Casein)
- 3 g Malt Extract
- 340 g Sucrose
- ad 1 L water
- DSMZ medium 65 GYM streptomyces medium
- 4 g Glucose
- 4 g Yeast extract
- 10 g Malt extract
- (2 g CaCO3 for plates)
- (12 g Agar for plates)
- ad 1 L water
Results and Discussion
BAP1+pMM64+pMM65 eventually produced indigoidine. We will now focus on assembling our own constructs starting with
pKH1. Streptomyces cultures look like yeast and have a strong smell as in the forest.
 "
"