Team:Heidelberg/Templates/DelH week 3
From 2013.igem.org
m |
|||
| (5 intermediate revisions not shown) | |||
| Line 53: | Line 53: | ||
<br/> | <br/> | ||
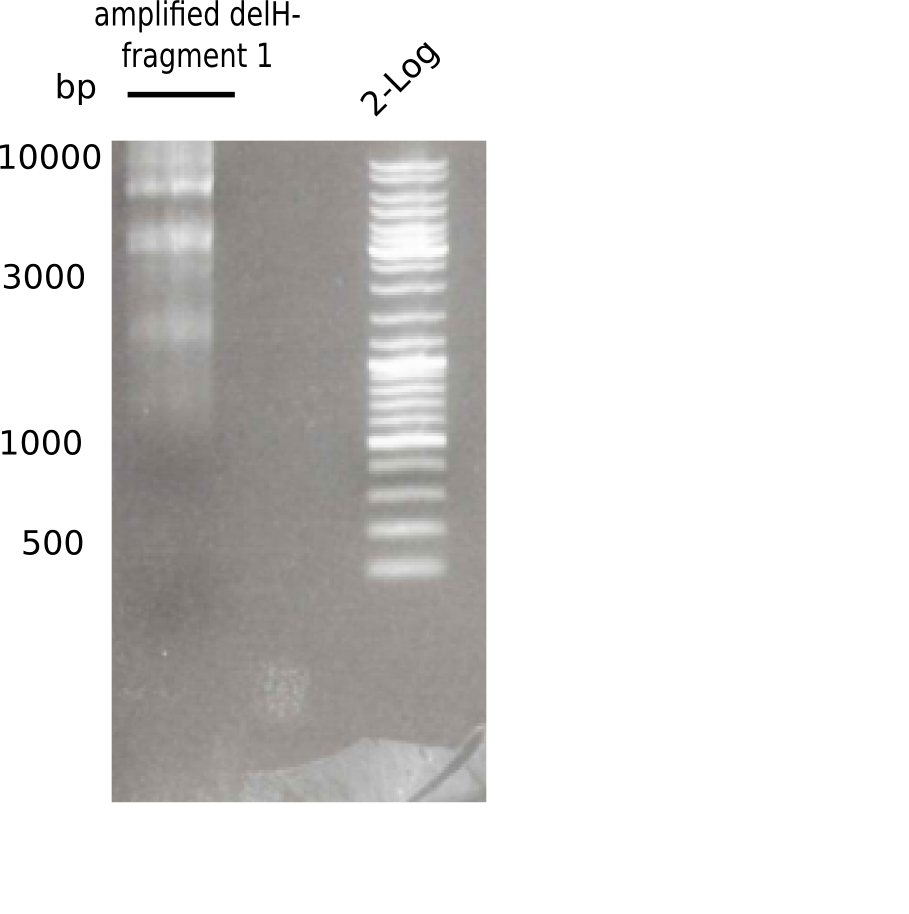
[[File:Heidelberg_20130513 2log F1.png|200px|thumb|right|'''Fig.3.1''' PCR of DelH fragment 1 (loaded 20 µL) <br> ''l1:''fragment 1 of DelH, ''l2:'' 2log ladder ]] | [[File:Heidelberg_20130513 2log F1.png|200px|thumb|right|'''Fig.3.1''' PCR of DelH fragment 1 (loaded 20 µL) <br> ''l1:''fragment 1 of DelH, ''l2:'' 2log ladder ]] | ||
| - | + | ||
Different unspecific bands were observed. | Different unspecific bands were observed. | ||
:=> Test digest to confirm identity. | :=> Test digest to confirm identity. | ||
<br/> | <br/> | ||
| - | + | <div style="clear:both"></div> | |
====Test Restriction Digest==== | ====Test Restriction Digest==== | ||
| Line 112: | Line 112: | ||
<br/> | <br/> | ||
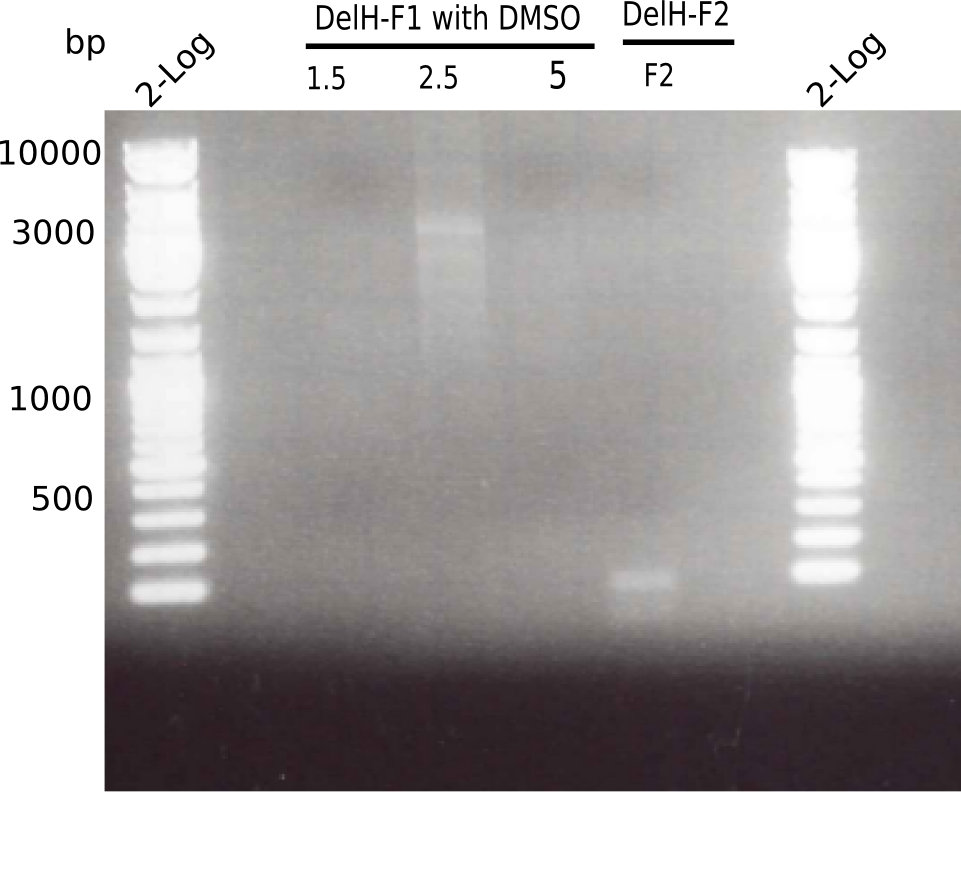
[[File:Heidelberg_20130516 2log 3xF1 F2.png|200px|thumb|right|'''Fig.3.2''' PCR of DelH fragment 1 (loaded 20 µL) <br> ''l1:''2log ladder, ''l2-4:'' fragment 1 of DelH, ''l5:'' fragment 2 of DelH ]] | [[File:Heidelberg_20130516 2log 3xF1 F2.png|200px|thumb|right|'''Fig.3.2''' PCR of DelH fragment 1 (loaded 20 µL) <br> ''l1:''2log ladder, ''l2-4:'' fragment 1 of DelH, ''l5:'' fragment 2 of DelH ]] | ||
| - | + | ||
There is only an unspecific band at the second PCR of F1 with 2.5 µl DMSO. | There is only an unspecific band at the second PCR of F1 with 2.5 µl DMSO. | ||
:=> Repeat using DMSO and altered PCR program. | :=> Repeat using DMSO and altered PCR program. | ||
<br/> | <br/> | ||
| + | <div style="clear:both"></div> | ||
====PCR Conditions F1.W3.C==== | ====PCR Conditions F1.W3.C==== | ||
{| class="wikitable" style="float:left; margin-right:1em" | {| class="wikitable" style="float:left; margin-right:1em" | ||
| Line 157: | Line 158: | ||
<br/> | <br/> | ||
[[File:Heidelberg_20130516 2log F1 F2 differentcycler.png|200px|thumb|right|'''Fig.3.3''' PCR of DelH fragment 1 (loaded 20 µL) <br> ''l1:''2log ladder, ''l2:'' fragment 1 of DelH, ''l3:'' fragment 2 of DelH, ''l4:'' fragment 1 of DelH, ''l5:'' fragment 2 of DelH ]] | [[File:Heidelberg_20130516 2log F1 F2 differentcycler.png|200px|thumb|right|'''Fig.3.3''' PCR of DelH fragment 1 (loaded 20 µL) <br> ''l1:''2log ladder, ''l2:'' fragment 1 of DelH, ''l3:'' fragment 2 of DelH, ''l4:'' fragment 1 of DelH, ''l5:'' fragment 2 of DelH ]] | ||
| - | + | ||
There was no band visible. | There was no band visible. | ||
:=> Repeat using DMSO and altered PCR program. | :=> Repeat using DMSO and altered PCR program. | ||
<br/> | <br/> | ||
| - | + | <div style="clear:both"></div> | |
===Amplification of DelH F2=== | ===Amplification of DelH F2=== | ||
====Gel Purification==== | ====Gel Purification==== | ||
| Line 214: | Line 215: | ||
<br/> | <br/> | ||
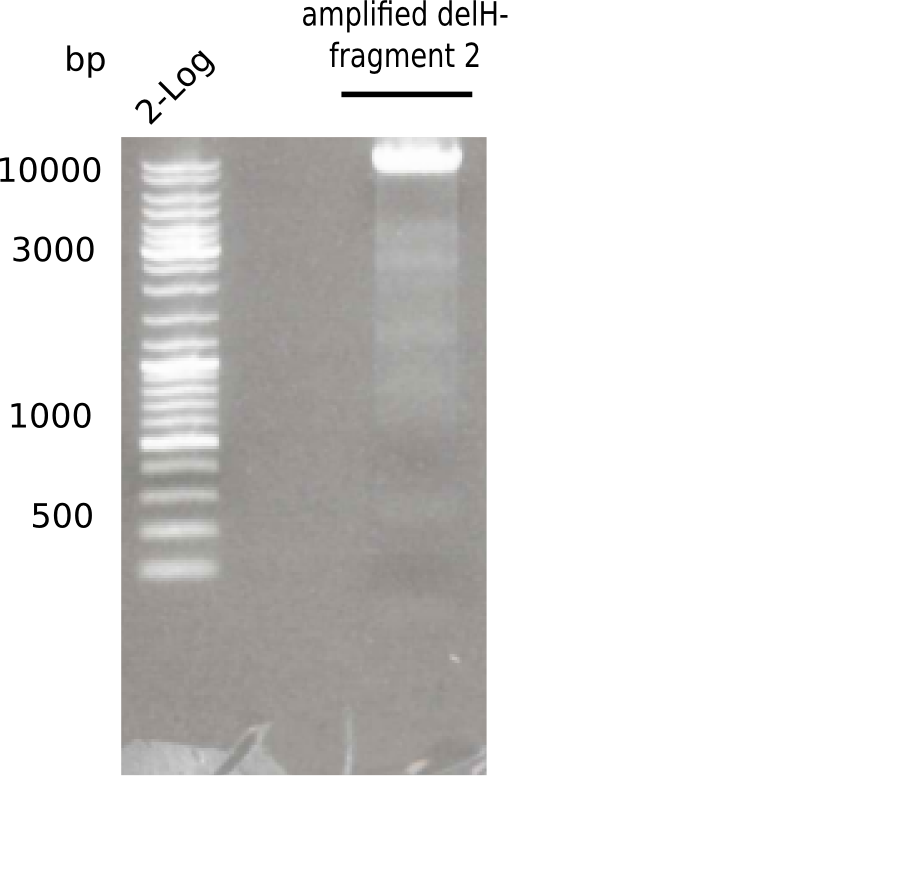
[[File:Heidelberg_20130513 2log F2.png|200px|thumb|right|'''Fig.3.4''' PCR of DelH fragment 1 (loaded 20 µL) <br> ''l1:''2log ladder, ''l2:'' fragment 2 of DelH ]] | [[File:Heidelberg_20130513 2log F2.png|200px|thumb|right|'''Fig.3.4''' PCR of DelH fragment 1 (loaded 20 µL) <br> ''l1:''2log ladder, ''l2:'' fragment 2 of DelH ]] | ||
| - | + | ||
Specific band was observed. | Specific band was observed. | ||
:=> Test digest to confirm identity. | :=> Test digest to confirm identity. | ||
<br/> | <br/> | ||
| - | + | <div style="clear:both"></div> | |
====Test Restriction Digest==== | ====Test Restriction Digest==== | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 271: | Line 272: | ||
<br/> | <br/> | ||
[[File:Heidelberg_20130516 2log 3xF1 F2.png|200px|thumb|right|'''Fig.3.2''' PCR of DelH fragment 1 (loaded 20 µL) <br> ''l1:''2log ladder, ''l2:'' fragment 1 of DelH, ''l3:'' fragment 2 of DelH ]] | [[File:Heidelberg_20130516 2log 3xF1 F2.png|200px|thumb|right|'''Fig.3.2''' PCR of DelH fragment 1 (loaded 20 µL) <br> ''l1:''2log ladder, ''l2:'' fragment 1 of DelH, ''l3:'' fragment 2 of DelH ]] | ||
| - | + | ||
There is no band visible. | There is no band visible. | ||
:=> Repeat using altered PCR program. | :=> Repeat using altered PCR program. | ||
<br/> | <br/> | ||
| - | + | <div style="clear:both"></div> | |
====PCR Conditions F2.W3.B==== | ====PCR Conditions F2.W3.B==== | ||
{| class="wikitable" style="float:left; margin-right:1em" | {| class="wikitable" style="float:left; margin-right:1em" | ||
| Line 316: | Line 317: | ||
<br/> | <br/> | ||
[[File:Heidelberg_20130516 2log F1 F2 differentcycler.png|200px|thumb|right|'''Fig.3.3''' PCR of DelH fragment 1 (loaded 20 µL) <br> ''l1:''2log ladder, ''l2:'' fragment 1 of DelH, ''l3:'' fragment 2 of DelH ]] | [[File:Heidelberg_20130516 2log F1 F2 differentcycler.png|200px|thumb|right|'''Fig.3.3''' PCR of DelH fragment 1 (loaded 20 µL) <br> ''l1:''2log ladder, ''l2:'' fragment 1 of DelH, ''l3:'' fragment 2 of DelH ]] | ||
| - | + | ||
There was a specific band at 8 Kb | There was a specific band at 8 Kb | ||
:=> Fragment was cut and gel extracted. | :=> Fragment was cut and gel extracted. | ||
<br/> | <br/> | ||
| - | + | <div style="clear:both"></div> | |
===Generation of Backbone pSB6A1-AraC-lacZ=== | ===Generation of Backbone pSB6A1-AraC-lacZ=== | ||
====Miniprep of Amplified Parts==== | ====Miniprep of Amplified Parts==== | ||
| Line 490: | Line 491: | ||
<br/> | <br/> | ||
[[File:Heidelberg_20130515 2log AraC-lacZ-pSB1C3.png|200px|thumb|right|'''Fig.3.5''' PCR of BB psB1C3-AraC-lacZ(loaded 20 µL) <br> ''l1:''2log ladder, ''l2:'' psB1C3-AraC-lacZ <br/> no yield ]] | [[File:Heidelberg_20130515 2log AraC-lacZ-pSB1C3.png|200px|thumb|right|'''Fig.3.5''' PCR of BB psB1C3-AraC-lacZ(loaded 20 µL) <br> ''l1:''2log ladder, ''l2:'' psB1C3-AraC-lacZ <br/> no yield ]] | ||
| - | + | ||
There was no fragments visible. | There was no fragments visible. | ||
:=> Is picked coloniy negative or did entire ligation not work out? | :=> Is picked coloniy negative or did entire ligation not work out? | ||
<br/> | <br/> | ||
| + | <div style="clear:both"></div> | ||
Latest revision as of 11:37, 23 October 2013
13-05 - 19-05-13
Amplification of DelH F1
Gel Purification
45 µl of DelH F1 PCR (08-05) product were run on a 0,8% agarose gel (1 h with 135V). Gel extraction was performed using gel extraction kit of Qiagen, and diluted in 25 µl ddH2O.
Result
| Sample | Concentration [ng/µl] |
|---|---|
| DelH F1 | 7 |
- => Because the concentration is so low, a re-PCR of the PCR product (08-05) is going to be done.
re-PCR Conditions F1.W3.A
| Reagent | DelH F1 |
|---|---|
| Expected length [Kb] | 10 |
| Template | 1 µl 1:10 gel purified F1 |
| Primer 10 µM fw | 0.5 µl DelH_f1_PacI_fw |
| Primer 10 µM rev | 0.5 µl DelH_f1_SalI_rev |
| Phusion Master Mix (2x) | 25 µl |
| DMSO | 2.5 µl |
| ddH2O | 20.5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 3:30 min | |
| 72 | 3:30 min | |
| 1 | 4 | inf |
Result
Expected length: 10 Kb
Different unspecific bands were observed.
- => Test digest to confirm identity.
Test Restriction Digest
| PCR product of DelH-F1 | BSA | NEBuffer 4 | Enzymes | ddH2O | Total amount |
|---|---|---|---|---|---|
| 18 µl | 5 µl | 5 µl | 2x 1.5 µl SalI-HF & PacI | 19 µl | 50 µl |
Afterwards, purification with the nucleotide removal kit was performed, but due to using wrong column, there was no product left.
- => Repeat amplification of DelH F1.
PCR Conditions F1.W3.B
| Reagent | DelH F1 | DelH F1 | DelH F1 |
|---|---|---|---|
| Expected length [Kb] | 10 | 10 | 10 |
| Template | Picked colony | Picked colony | Picked colony |
| Primer 10 µM fw | 0.5 µl DelH_f1_PacI_fw | 0.5 µl DelH_f1_PacI_fw | 0.5 µl DelH_f1_PacI_fw |
| Primer 10 µM rev | 0.5 µl DelH_f1_SalI_rev | 0.5 µl DelH_f1_SalI_rev | 0.5 µl DelH_f1_SalI_rev |
| Phusion Master Mix (2x) | 25 µl | 25 µl | 25 µl |
| DMSO | 1.5 µl | 2.5 µl | 5 µl |
| ddH2O | 21.5 µl | 20.5 µl | 18 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 s |
| 66 | 5 s | |
| 72 | 3:30 min | |
| 1 | 72 | 3:30 min |
| 1 | 4 | inf |
Result
Expected length: 10 Kb
There is only an unspecific band at the second PCR of F1 with 2.5 µl DMSO.
- => Repeat using DMSO and altered PCR program.
PCR Conditions F1.W3.C
| Reagent | DelH F1 | DelH F1 |
|---|---|---|
| Expected length [Kb] | 10 | 10 |
| Template | 1 µl glycerol stock | 1 µl glycerol stock |
| Primer 10 µM fw | 0.5 µl DelH_f1_PacI_fw | 0.5 µl DelH_f1_PacI_fw |
| Primer 10 µM rev | 0.5 µl DelH_f1_SalI_rev | 0.5 µl DelH_f1_SalI_rev |
| Phusion Master Mix (2x) | 25 µl | 25 µl |
| DMSO | 2.5 µl | 2.5 µl |
| ddH2O | 20.5 µl | 20.5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 s |
| 66 | 5 s | |
| 72 | 3:30 min | |
| 1 | 72 | 3:30 min |
| 1 | 4 | inf |
Result
Expected length: 10 Kb
There was no band visible.
- => Repeat using DMSO and altered PCR program.
Amplification of DelH F2
Gel Purification
45 µl of DelH F1 PCR (07-05) product were run on a 0,8% agarose gel (1 h with 135V). Gel extraction was performed using gel extraction kit of Qiagen, and diluted in 25 µl ddH2O.
Result
| Sample | Concentration [ng/µl] |
|---|---|
| DelH F2 | 6 |
- => Because the concentration is so low, a re-PCR of the PCR product (07-05) is going to be done.
Re-PCR Conditions F2.W3.A
| Reagent | DelH-fragment2 |
|---|---|
| Expected length [Kb] | 8 |
| Template | 1 µl 1:10 gel purified F2 |
| Primer 10 µM fw | 0.5 µl DelH_f2_SalI_fw |
| Primer 10 µM rev | 0.5 µl DelH_f2_KpnI_rev |
| Phusion Master Mix (2x) | 25 µl |
| DMSO | 2.5 µl |
| ddH2O | 20.5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 s |
| 66 | 5 s | |
| 72 | 3:30 min | |
| 72 | 3:30 min | |
| 1 | 4 | inf |
Result
Expected length: 8 Kb
Specific band was observed.
- => Test digest to confirm identity.
Test Restriction Digest
| PCR product of DelH-F1 | BSA | NEBuffer 4 | Enzymes | ddH2O | Total amount |
|---|---|---|---|---|---|
| 18 µl | 5 µl | 5 µl | 2x 1.5 µl SalI-HF & KpnI-HF | 19 µl | 50 µl |
Afterwards, a purification with the nucleotide removal kit was performed, but due to using wrong column, there was no product left.
- => Repeat amplification of DelH F2.
PCR Conditions F2.W3.B
| Reagent | DelH F2 |
|---|---|
| Expected length [Kb] | 8 |
| Template | Picked colony |
| Primer 10 µM fw | 0.5 µl DelH_f2_SalI_fw |
| Primer 10 µM rev | 0.5 µl DelH_f2_KpnI_rev |
| Phusion Master Mix (2x) | 25 µl |
| DMSO | 0 µl |
| ddH2O | 23 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:45 min | |
| 1 | 72 | 2:45 min |
| 1 | 4 | inf |
Result
Expected length: 8 Kb
There is no band visible.
- => Repeat using altered PCR program.
PCR Conditions F2.W3.B
| Reagent | DelH F2 |
|---|---|
| Expected length [Kb] | 8 |
| Template | 1 µl glycerol stock |
| Primer 10 µM fw | 0.5 µl DelH_f2_SalI_fw |
| Primer 10 µM rev | 0.5 µl DelH_f2_KpnI_rev |
| Phusion Master Mix (2x) | 25 µl |
| DMSO | 0 µl |
| ddH2O | 23 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:45 min | |
| 1 | 72 | 2:45 min |
| 1 | 4 | inf |
Result
Expected length: 8 Kb
There was a specific band at 8 Kb
- => Fragment was cut and gel extracted.
Generation of Backbone pSB6A1-AraC-lacZ
Miniprep of Amplified Parts
DH10ß containing I13453 and K20600 (for AraC) and backbone pSB1AK3 (lacZ) were grown ON at 37°C and minipreped.
Result
| Sample | Concentration [ng/µl] |
|---|---|
| lacZ | 42.5 |
| AraC (5) | 26.5 |
| AraC (6) | 34.5 |
Restriction Digest
- AraC was cut with EcoRI-HF and SpeI buffered in NEB4
- LacZ was cut with XbaI and PstI buffered in NEB3
| Reagent | Amount [µl] |
|---|---|
| DNA | 22.5 |
| Enzymes | 1.5 each |
| Buffer (10x) | 5 |
| BSA (10x) | 5 |
| ddH2O | 14.5 |
- Incubated for 1h at 37°C
- Purified with Qiagen Kit and diluted in 20 µl ddH2O
Result
| Sample | Concentration [ng/µl] |
|---|---|
| lacZ | 33.5 |
| AraC (5) | 19.5 |
| AraC (6) | 25.5 |
Ligation of pSB6A1, AraC and lacZ
| Reagent | Amount [µl] |
|---|---|
| pSB6A1 | 1.5 |
| lacZ | 4 |
| AraC | 3 |
| Ligase | 1.5 |
| Buffer | 2 |
| ddH2O | 8.5 |
- Incubated at RT for 45 min
- Chemical transformation of competent DH10ß
- Streaked on LB Amp plates
- Incubation ON at 37°C
Miniprep of pSB6A1-AraC-lacZ
- One colony each was picked and grown in 2 ml LB Amp ON
- Cultures were minipreped.
Result
| Sample | Concentration [ng/µl] |
|---|---|
| pSB6A1-AraC-lacZ (5) | 57 |
| pSB6A1-AraC-lacZ (6) | 73 |
Restriction Digest of Backbone pSB6A1-AraC-lacZ
- AraC-lacZ was cut from pSB6A1-AraC-lacZ using PstI & EcoRI-HF
| Reagent | Amount [µl] |
|---|---|
| Miniprep DNA | 10 |
| Enzymes | 1.5 each |
| Buffer NEB 2(10x) | 5 |
| BSA (10x) | 5 |
| ddH2O | 27 |
- Incubated for 1 h at 37°C
- Purified with Qiagen kit and diluted in 20 µl ddH2O
Result
| Sample | Concentration [ng/µl] |
|---|---|
| pSB6A1-AraC-lacZ (5) D+P | 18 |
| pSB6A1-AraC-lacZ (6) D+P | 26 |
Ligation of AraC-lacZ with pSB1C3
| Reagent | Amount [µl] |
|---|---|
| DNA of pSB1C3 | 10 |
| DNA of AraC-lacZ (6) | 5 |
| Ligase | 1 |
| Buffer | 2 |
| ddH2O | 2 |
- Incubated at RT for 50 min
- Heat inactivation at 70°C for 5 min
- 10 µl were used for a chemical transformation in TOP10 and plated on LB Chlor
- Incubation ON at 37°C
Conlony-PCR Conditions BB.W3.A
| Reagent | psB1C3-AraC-lacZ (5) |
|---|---|
| Expected length [Kb] | ? |
| Template | Picked colony of psB1C3-AraC-lacZ (5) |
| Primer 10 µM fw | 0.5 µl |
| Primer 10 µM rev | 0.5 µl |
| Phusion Master Mix (2x) | 25 µl |
| DMSO | - |
| ddH2O | 24 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 1:30 min | |
| 1 | 72 | 1:30 min |
| 1 | 4 | inf |
Result
Expected length:
There was no fragments visible.
- => Is picked coloniy negative or did entire ligation not work out?
 "
"