Team:Braunschweig/Project/Description
From 2013.igem.org
| (2 intermediate revisions not shown) | |||
| Line 32: | Line 32: | ||
<img alt="linie rot 8pix hoch" src="https://static.igem.org/mediawiki/2013/0/07/Team_Braunschweig_Red_line.jpg" width="850" height="1" /></p> | <img alt="linie rot 8pix hoch" src="https://static.igem.org/mediawiki/2013/0/07/Team_Braunschweig_Red_line.jpg" width="850" height="1" /></p> | ||
| - | <img alt="Project scheme" src="https://static.igem.org/mediawiki/2013/6/64/Braunschweig_Project.png" width="450" align="right" vspace="10" hspace="20" /><p style="text-align: justify">In our project, we engineer three different E. coli strains to grow in | + | <a href="https://static.igem.org/mediawiki/2013/6/64/Braunschweig_Project.png"><img alt="Project scheme" src="https://static.igem.org/mediawiki/2013/6/64/Braunschweig_Project.png" width="450" align="right" vspace="10" hspace="20" /></a><p style="text-align: justify">In our project, we engineer three different <i>E. coli</i> strains to grow in |
a consortium. The survivability of the bacteria is coupled to a Quorum | a consortium. The survivability of the bacteria is coupled to a Quorum | ||
Sensing (QS) dependent antibiotic resistance.</p> | Sensing (QS) dependent antibiotic resistance.</p> | ||
| Line 65: | Line 65: | ||
<h1>Our sponsors</h1></p> | <h1>Our sponsors</h1></p> | ||
<img alt="linie rot 8pix hoch" src="https://static.igem.org/mediawiki/2013/0/07/Team_Braunschweig_Red_line.jpg" width="850" height="1" /></p> | <img alt="linie rot 8pix hoch" src="https://static.igem.org/mediawiki/2013/0/07/Team_Braunschweig_Red_line.jpg" width="850" height="1" /></p> | ||
| - | <img src="https://static.igem.org/mediawiki/2013/ | + | <img src="https://static.igem.org/mediawiki/2013/9/9e/SponsorenBS.png" width="850px" /></p> |
</div> | </div> | ||
</html> | </html> | ||
Latest revision as of 19:58, 27 October 2013

Our project
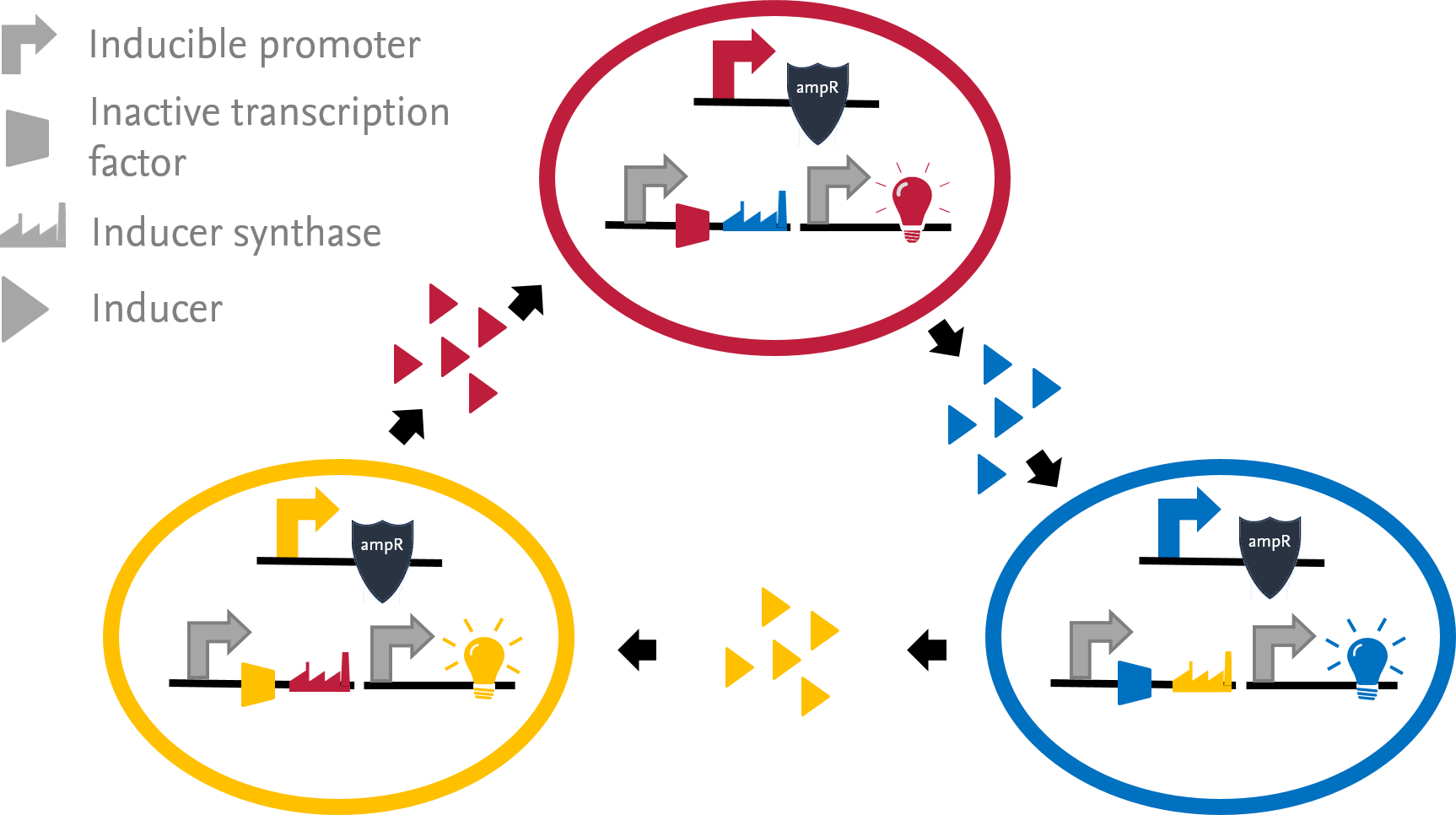
In our project, we engineer three different E. coli strains to grow in a consortium. The survivability of the bacteria is coupled to a Quorum Sensing (QS) dependent antibiotic resistance.
Each strain maintains a constitutive expression of an inactive transcription activator (LuxR, LasR or RhlR). Inducers are synthesized by different synthases (LuxI, LasI or RhlI) that are each expressed in one strain and subsequently secreted into the medium. Once taken up by a cell, the inducers bind to the corresponding, inactive transcription factors to render them functional. As a result, the antibiotic resistance under the control of an inducible promoter is expressed.
To measure the culture composition, we include a strain specific chromoprotein as reporter. As an individual strain can only grow if the inactive transcription factor is activated, the three strains are forced into a symbiotic dependency. If one strain dies, the whole consortium dies.
The benefit of bacterial consortia for synthetic biology is the ability to perform very complex tasks. In applications like wastewater treatment, soil remediation or the complicated synthesis of biotechnological products, the whole reaction can be split into small reactions to share the task among the different strains. Also, a self-regulating bacterial culture with intra-consortial dependencies offers great advances in biosafety. To shut down the whole bacterial consortium, only one strain has to be eliminated.
Our sponsors

 "
"
